Quantum microRNA Assessment of COVID-19 RNA Vaccine: Hidden Potency of BNT162b2 SASR-CoV-2 Spike RNA as MicroRNA Vaccine
Or "would you like rice with that?"
Any extracts used in the following article are for non commercial research and educational purposes only and may be subject to copyright from their respective owners.
Also see follow up post including a walkthrough of the Crimson paper:
Hat tip to @Arkmedic, Dr. Ah Khan Syed:
#miRNAgate - what the hell have they unleashed?
Quantum microRNA Assessment of COVID-19 RNA Vaccine: Hidden Potency of BNT162b2 SASR-CoV-2 Spike RNA as MicroRNA Vaccine (2021)
Abstract
Objective: The pandemic of coronavirus disease 2019 (COVID-19) is caused by infection with severe respiratory syndrome human coronavirus 2 (SARS-CoV-2). The spike (S) RNA of SARS-CoV-2 is used to build the COVID-19 vaccine. Although the level of immune response elicited by the full-length S RNA vaccine and the Receptor Binding Domain (RBD) vaccine that is a part of S is similar, the full-length S RNA vaccine is safer and lower reactogenicity than the RBD vaccine. However, the reason has not yet been clarified. On the other hand, the COVID-19 RNA vaccine with the S sequences may produce viral microRNAs (miRNAs). But there are no miRNA assessments for the safety of the COVID-19 vaccines.
Therefore, evaluation as miRNA vaccines is necessary for risk management of vaccination.
Materials and Methods: miRNA Fold was used for pre-miRNA prediction in SARS-CoV-2 S RNA. MR-micro-T was used for protein target search. The integrated network algorithm as the miRNA entangling sorter (METS) analysis by using quantum miRNA language plus Artificial Intelligence (AI) were used as previously described.
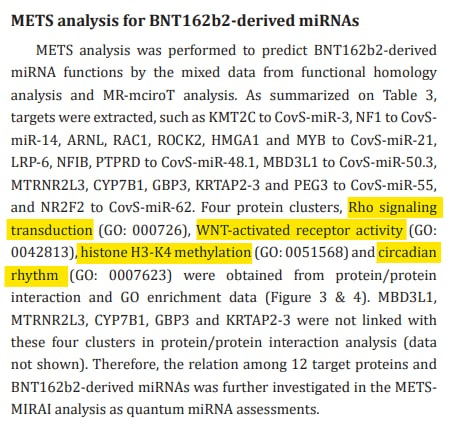
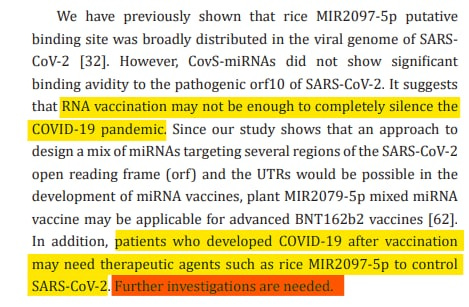
Results: In computation, sixteen SARS-CoV-2-S-derived miRNAs bound to the negative strand S RNA with quite strong avidities. Further, CovS-miR-21 downregulated Rho associated coiled-coil containing protein kinase (ROCK2) and aryl hydrocarbon receptor nuclear translocator like (ARNTL), and CovS-miR-3 decreased lysine methyltransferase 2C (KMT2C). Therefore, in the METS analysis, CovS-miR-21 suppressed the function of Ras homolog family member A (RhoA)/Rock2 signaling and circadian rhythm, and CovS-miR-3 inhibited histone H3-K4 methylation.
Conclusion: We found that BNT162b2 inhibits SARS-CoV-2 replication through degradation of negative strand viral RNAs that are completely paired with SARS-CoV-2 S-derived miRNAs. Further, CovS-miR-21 derived from BNT162b2 restores circadian rhythm and attenuate immunogenicity. Quantum miRNA assessments showed that the BNT162b2 RNA vaccine has a character of miRNA vaccine and is an excellent vaccine with high efficacy and low side-effects.
Keywords: COVID-19; SARS-CoV-2; BNT162b2; RNA vaccine; microRNA; Computer simulation; In silico; Circadian rhythm; Rho pathway
https://crimsonpublishers.com/aics/pdf/AICS.000552.pdf
In silico prediction of human genes as potential targets for rice miRNAs (2020)
Highlights
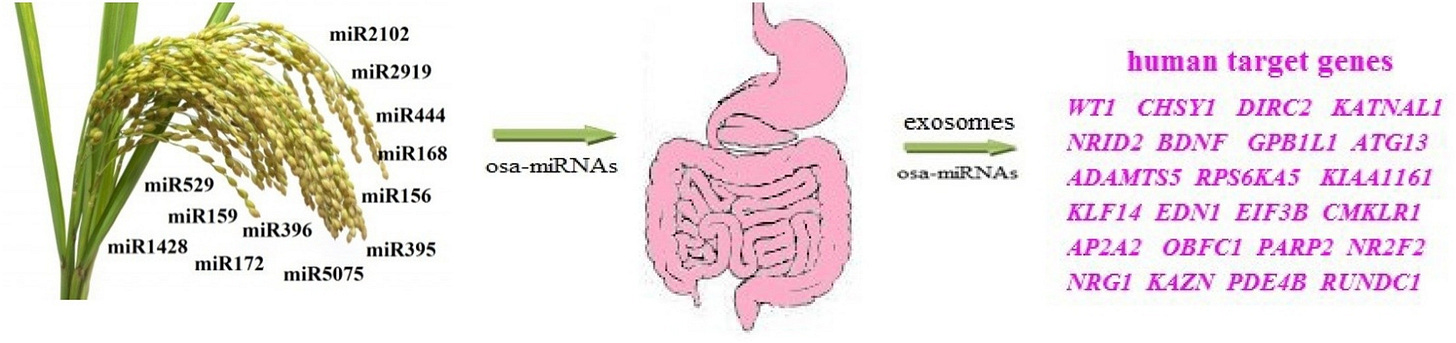
•A highly complementary interaction of single and family osa-miRNAs with mRNAs of human genes was shown.
•The nucleotides of the binding sites of many osa-miRNAs in mRNA of the target genes are conservative compared to the flanking nucleotides.
•Most osa-miRNAs can affect target mRNA human genes involved in the development of some human diseases.
"Conclusion
Target mRNA human genes of osa-miRNAs are also candidate genes of cancer, cardiovascular and neurodegenerative diseases."
"The effect of miRNAs can be both positive, contributing to the cure of diseases, and negative, causing a wide range of diseases.”
https://www.sciencedirect.com/science/article/pii/S1476927120301420
This patent was filed in 2015. It reads like a snake oil salesman's pitch of a cure-all for everything afflicting mankind. Just silence or upregulate the relevant gene, job done:
Patent application title: Methods and Compositions to Modulate RNA Processing Inventors: Hannele Ruohoa-Baker (Seattle, WA, US) Pratyusha Banik (Bothell, WA, US) Alan Beem (Seattle, WA, US) Sandra Shannon (Sammamish, WA, US) Henrik Sperber (Seattle, WA, US) IPC8 Class: AC12N1585FI USPC Class: 514 44 R Class name: Publication date: 2015-11-26 Patent application number: 20150337332
Abstract: The present disclosure provides methods and compositions to selectively modulate RNA processing. The methods and compositions selectively enhance or repress RNA processing by up- or down-regulating Drosha expression and/or by providing RNA sequences with mis-matches introduced or removed 5 and/or 9-12 nucleotide positions from the Drosha cutting site. Therapeutic uses of the methods and compositions are also described.
[0099] miRNA, artificial miRNA and Drosha up- and down-regulation disclosed herein can be used in therapeutic applications directed to infectious diseases, i.e., those arising from the presence of a foreign microorganism in the body. A microbial antigen, as used herein, is an antigen of a microorganism. Microorganisms include but are not limited to, infectious viruses, infectious bacteria, and infectious fungi.
More: