Spike protein (inc vax) induced immunodeficiency & carcinogenesis megathread #23: High-content screening of coronavirus genes for innate immune suppression reveals enhanced potency of SARS-CoV-2
(preprint, 2021)
Any extracts used in the following article are for non commercial research and educational purposes only and may be subject to copyright from their respective owners.
Background and hat tips:
part II: Golden Silkworms in Pandora's Box
Why understanding COVID-19 and Seasonal Influenza as Quasispecies Mutant Swarms reveals the Quantum Origins and Cryptic Fates of Human Pandemics.
Harvard2TheBigHouse
Apr 1, 2021
https://harvard2thebighouse.substack.com/p/part-ii-understanding-covid-19-and?s=r
The paper:
High-content screening of coronavirus genes for innate immune suppression reveals enhanced potency of SARS-CoV-2 proteins
Summary
Suppression of the host intracellular innate immune system is an essential aspect of viral replication. Here, we developed a suite of medium-throughput high-content cell-based assays to reveal the effect of individual coronavirus proteins on antiviral innate immune pathways. Using these assays, we screened the 196 protein products of seven coronaviruses (SARS-CoV-2, SARS-CoV-1, 229E, NL63, OC43, HKU1 and MERS). This includes a previously unidentified gene in SARS-CoV-2 encoded within the Spike gene. We observe immune-suppressing activity in both known host-suppressing genes (e.g., NSP1, Orf6, NSP3, and NSP5) as well as other coronavirus genes, including the newly identified SARS-CoV-2 protein. Moreover, the genes encoded by SARS-CoV-2 are generally more potent immune suppressors than their homologues from the other coronaviruses. This suite of pathway-based and mechanism-agnostic assays could serve as the basis for rapid in vitro prediction of the pathogenicity of novel viruses based on provision of sequence information alone.
Figure 3.
Inhibition of IRF-3, NFkB and STAT1 active nuclear levels by coronavirus genes.
A-C. Statistically significant average inhibition scores from two to five experiments for each gene from each virus (e.g., Figure 2H) were combined. Top: Vertical bar graphs were generated by averaging the inhibition scores for a given gene across all viruses with that gene. Left: Horizontal bar graphs show the sum of all of the inhibition scores of genes from a given virus. Bottom: Heat map of average scores across all relevant stimuli in all experiments (each gene for each virus). Red indicates inhibition of innate immune signaling, green shows enhancement; white indicates a given gene is not present in a given virus; gray indicates a gene not tested. A. Inhibition of IRF-3 translocation and enhancement of degradation in cells stimulated by cGAMP plus low molecular weight polyI:C in lipofectamine plus free high molecular weight polyI:C. B. Inhibition of NFkB translocation and enhancement of degradation in cells stimulated by LMW-polyI:C in lipofectamine plus free HMW polyI:C. C. Inhibition of STAT1 translocation, enhancement of degradation, and formation of phospho-STAT1 in cells stimulated by interferon-alpha. See Supplementary Information for full details.
Discussion
In this work, we characterized the ability of the proteins encoded by seven coronaviruses to suppress host intracellular innate immune signaling pathways using high-content, medium-throughput cell-based assays. The first assay examined the nuclear localization of the transcription factors IRF-3, NFkB, and STAT1, which play key roles in the elaboration of the Type I interferon response to viral infection, and which are major targets for inhibition by other viruses (Figure 2; Hoffmann et al., 2015). Viruses act on these pathways by a variety of mechanisms, such as specific inhibition of upstream signal transduction proteins, general inhibition of nuclear import, and enhancement of degradation. Our assays involved transfection of a non-transformed cell line with expression vectors encoding each gene followed by addition of a stimulator of innate immune signaling, and then immunofluorescence staining and automated image processing. We found that:
Localization of IRF-3 and NFkB to the nucleus was inhibited by a number of coronavirus genes, including known innate immune suppressors such as NSP5, NSP3, Orf7a and NSP1 (Freitas et al., 2020; Lei et al., 2018), and proteins not previously known to have such an activity, including CaORF15 and NSP9.
Two genes were found to have an impact on STAT1 activation: NSP1 and Orf6. Other groups previously found that that NSP1 binds to the ribosome and inhibits translation of a subset of host proteins (Tidu et al., 2021), and that Orf6 protein inhibits the nuclear protein import factor Nup98 (Frieman et al., 2007). Our results are consistent with these observations.
In the 3’ region of the coronavirus genome, which encodes accessory proteins that vary among the coronaviruses, SARS-CoV-2 proteins are more potent in their inhibition of innate immune signaling than the corresponding SARS-CoV-1 protein (Figure 3).
SARS-CoV-2 appears to encode a novel protein (“CaORF15”), which is predicted to have three transmembrane domains. CaORF15 is encoded in an alternate reading frame within the Spike coding sequence, and is not found in SARS-CoV-1 but is present in the more closely related bat virus RaTG13. CaORF15 inhibits IRF-3 nuclear accumulation particularly through the STING pathway (Figure 3, Supplemental Information) and also suppresses the innate immune system in the yeast-mammalian cell fusion assay (Figure 5).
When the inhibitory effects of coronavirus genes are examined in aggregate, SARS-CoV-2 appears to have more potential to suppress the innate immune system. This is based on summation of effects on IRF-3 and NFkB nuclear localization (Figure 3A and 3B), the strength of NSP1 inhibition of NFkB-mediated gene expression (Figure 4), and the genes showing effects in the yeast cell fusion assay (Figure 5).
Full paper:
https://www.biorxiv.org/content/10.1101/2021.03.02.433434v1
Interpretation:
SARS-CoV-2 proteins are potent suppressors of innate immunity (2021)
Most pathogenic viruses, such as the causative agent of the current coronavirus disease 2019 (COVID-19) pandemic, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), affect normal human biology to co-opt cellular metabolism and replicate successfully. One key pathway is the suppression of innate host immunity.
A new paper on the preprint bioRxiv* server describes the strong immune-suppressing activity of SARS-CoV-2 genes, among other coronavirus genes.
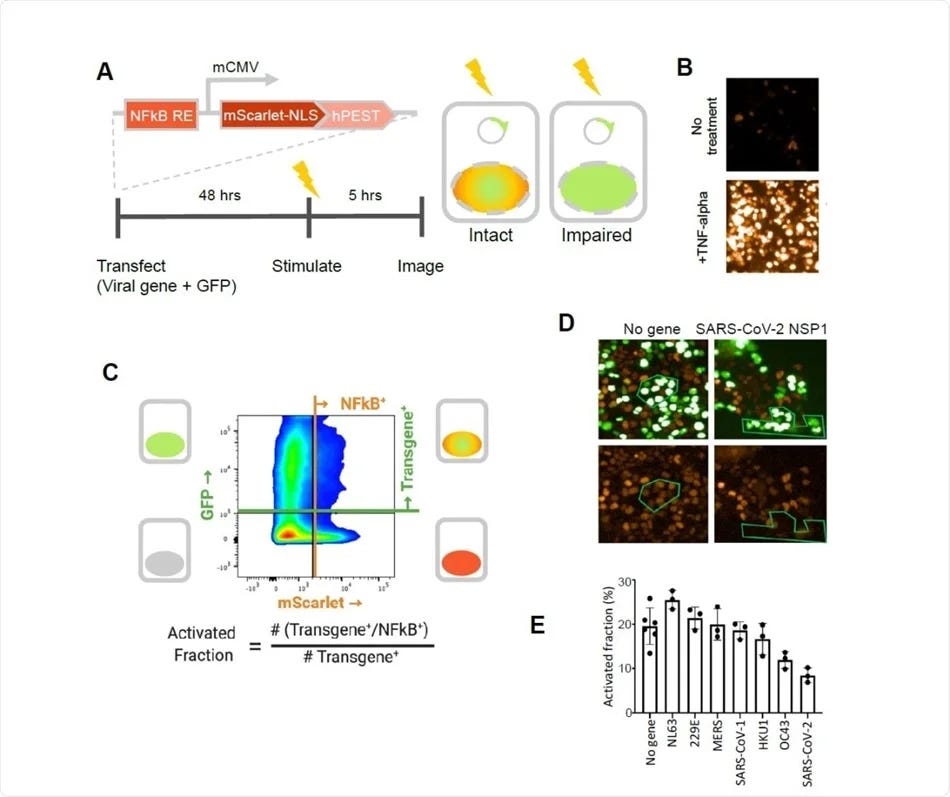
SARS-CoV-2 NSP1 strongly inhibits TNF-alpha activation of an NFkB reporter gene. A. A fluorescent NFkB reporter consisting of a 5x repeat of the NFkB consensus sequence (NFkB RE) upstream of a minimal CMV promoter (mCMV), and a human codon-optimized, nuclear localized mScarlet (mScarlet-NLS) fused to an hPEST degradation tag (hPEST) was constructed and stably integrated into HEK293 cells. Reporter HEK293T cells were co-transfected with expression plasmids encoding GFP and a virus gene; 48 hours later, 5 ng/ml TNF-? is added; and 5 hours later, red and green fluorescence was quantified. B. HEK293T cells stably transduced with the reporter responded to human TNF-? by expression of mScarlet. C. Flow cytometry analysis of transfected, TNF?-treated cells. Upper quadrants are transfected cells. The upper right quadrant represents double-positive cells that are transfected and also 338 express the inducible transgene. The activated fraction was calculated as the ratio of double positive cells to all GFP-positive cells. D. Reporter HEK293T cells were co-transfected with GFP and no-gene or SARS-CoV-2 NSP1 expression vectors, followed by TNF-? stimulation and imaging of red and green fluorescence. Green polygons highlight identical populations of cells in both images for representative comparisons. E. Activated fractions compared among NSP1s from several coronaviruses. Error bars represent the standard deviation of at least three technical replicates.
What are the implications?
This study shows the utility of a cell-based assay in reflecting the extent of viral replication in response to immune suppression.
Overall, SARS-CoV-2 proteins are strongly suppressive of the innate immune response compared to the average of all coronavirus genes put together, based on the following effects: IRF-3 and NFkB nuclear localization, NSP1 inhibition of NFkB-mediated gene expression, and effects in the yeast cell fusion assay.
In the latter, only the presence of active innate immune suppression yields active virus. Thus, the recovery of the virus indicates the presence of such viral proteins, in general, but if the host inhibition is very powerful, the virus may not be able to replicate at all. This appears to be the case with SARS-CoV-2 NSP1.
Importantly, the presence of the CaORF15 gene within the spike sequence means that it is part of many vaccines based on the wildtype spike, and as such, may hinder the generation of immunity in response to the vaccine. This is not the case with the Pfizer/BioNTech vaccine, which has a modification within this region.
The suppression of innate immune responses is more likely to be aligned with asymptomatic spread rather than increased disease severity.
More: