Original Antigenic Sin observed: "Ablation of CD8+ T cell recognition of an immunodominant epitope in SARS-CoV-2 Omicron variants BA.1, BA.2 and BA.3"
- A walkthrough of the paper with definitions
Substack limitations
Once Substacks’ support the use of internal hyperlinks I will add them to the abstract. Otherwise please use a “Find In Page” keyword search to navigate or click on the footnote text numbers.
If received via email I recommend clicking on the hyperlinked title to read the latest correctly formatted version in a browser. Unfortunately it is not possible to email out revised versions.
Coming soon: Therapeutic properties of Baicalin, A literature review, which is taking several weeks to complete due to the broad spectrum nature of its therapeutic properties and the number of signalling pathways that it interacts with, hence a large body of research which requires consideration. Dr Johanna Dienert is also very kindly contributing to the review.
This Substack is a walkthrough of the following paper, published 27th October 2022 by Swaminathan et al:1
Ablation of CD8+ T cell recognition of an immunodominant epitope in SARS-CoV-2 Omicron variants BA.1, BA.2 and BA.3.
Definitions
Original antigenic sin (OAS) is the phenomenon in which prior exposure to an antigen leads to a subsequent suboptimal immune response to a related antigen. Immune memory normally allows for an improved and rapid response to antigens previously seen and is the mechanism by which vaccination works.2
Ablated T cell activation in this study is the failure of previously reactive and immunodominant T cells to respond to mutated versions of SARS-CoV-2, otherwise known as original antigenic sin.
The authors may have considered it too controversial to include OAS in the title, but it may be a little misleading as ablation normally refers to selectively destroying or removing cells in an organism3, as in immunoablation prior to a stem cell transplant, for instance.
Also see:
Leukocyte: a type of blood cell that lacks hemoglobin and is therefore colorless. Leukocytes are larger in size and fewer in number than erythrocytes; normally the blood has about 8000 of them per mm3. In contrast to erythrocytes, leukocytes can move about under their own power with ameboid movement. Their chief functions are to act as scavengers and to help fight infections. Called also white cell or corpuscle and white blood cell or corpuscle. adj., adj leukocyt´ic.
Leukocytes may be classified in two main groups: the granular leukocytes are the basophils, eosinophils, and neutrophils, and the nongranular leukocytes are the lymphocytes and monocytes. About 63 per cent of all leukocytes are neutrophils; 2.5 per cent are eosinophils; and the remaining types constitute less than 1 per cent each.
Leukocytes are actively engaged in the destruction or neutralization of invading microorganisms and are quickly transported to the vicinity of infection or inflammation, so that they can move through the blood vessel wall to reach the site of injury. For this reason, their life span in the blood is usually very short. When infection is present their numbers are greatly increased and they also become more mobile and move back and forth between the blood, lymph, and tissues. The granulocytes and monocytes are phagocytic, swallowing or ingesting the foreign particles with which they come in contact. During the process of phagocytosis the phagocytes themselves are destroyed. The two types of lymphocytes involved in immunity are B lymphocytes (B cells), which play a role in humoral immunity, and T lymphocytes (T cells), which are important in cell-mediated immunity. Plasma cells are activated B cells that secrete antibodies. Monocytes are also involved in some immune processes.4
Peptides (from Ancient Greek πεπτός (peptós) 'digested', from πέσσειν (péssein) 'to digest') are short chains of amino acids linked by peptide bonds. Chains of fewer than twenty amino acids are called oligopeptides, and include dipeptides, tripeptides, and tetrapeptides.
A polypeptide is a longer, continuous, unbranched peptide chain. Hence, peptides fall under the broad chemical classes of biological polymers and oligomers, alongside nucleic acids, oligosaccharides, polysaccharides, and others.
A polypeptide that contains more than approximately 50 amino acids is known as a protein. Proteins consist of one or more polypeptides arranged in a biologically functional way, often bound to ligands such as coenzymes and cofactors, or to another protein or other macromolecule such as DNA or RNA, or to complex macromolecular assemblies.
Amino acids that have been incorporated into peptides are termed residues. A water molecule is released during formation of each amide bond. All peptides except cyclic peptides have an N-terminal (amine group) and C-terminal (carboxyl group) residue at the end of the peptide (as shown for the tetrapeptide in the image).

An epitope, also known as antigenic determinant, is the part of an antigen that is recognized by the immune system, specifically by antibodies, B cells, or T cells. The epitope is the specific piece of the antigen to which an antibody binds. The part of an antibody that binds to the epitope is called a paratope. Although epitopes are usually non-self proteins, sequences derived from the host that can be recognized (as in the case of autoimmune diseases) are also epitopes.5
HLA-A*29:02 is one of the two main subtypes of HLA-A29, the other being HLA-A*29:01. In the general population HLA-A*29:02 is most frequent in whites, while HLA-A*29:01 is more frequent in Asians.6
HLA-A29 (A29) is a human leukocyte antigen serotype within HLA-A serotype group. The serotype is determined by the antibody recognition of α29 subset of HLA-A α-chains. For A29, the alpha "A" chain are encoded by the HLA-A*29 allele group and the β-chain are encoded by B2M locus. This group currently is dominated by A*2902. A29 and A*29 are almost synonymous in meaning. A29 is a split antigen of the broad antigen serotype A19. A29 is a sister serotype of A30, A31, A32, A33, and A74.7
The human leukocyte antigen (HLA) system or complex is a complex of genes on chromosome 6 in humans which encode cell-surface proteins responsible for the regulation of the immune system. The HLA system is also known as the human version of the major histocompatibility complex (MHC) found in many animals.
HLAs corresponding to MHC class I (A, B, and C), all of which are the HLA Class1 group, present peptides from inside the cell. For example, if the cell is infected by a virus, the HLA system brings fragments of the virus to the surface of the cell so that the cell can be destroyed by the immune system. These peptides are produced from digested proteins that are broken down in the proteasomes. In general, these particular peptides are small polymers, of about 8-10 amino acids in length. Foreign antigens presented by MHC class I attract T-lymphocytes called killer T-cells (also referred to as CD8-positive or cytotoxic T-cells) that destroy cells.8
The major histocompatibility complex (MHC) is a large locus on vertebrate DNA containing a set of closely linked polymorphic genes that code for cell surface proteins essential for the adaptive immune system. These cell surface proteins are called MHC molecules.
This locus got its name because it was discovered via the study of transplanted tissue compatibility. Later studies revealed that tissue rejection due to incompatibility is only a facet of the full function of MHC molecules: binding an antigen derived from self-proteins, or from pathogens, and bringing the antigen presentation to the cell surface for recognition by the appropriate T-cells. MHC molecules mediate the interactions of leukocytes, also called white blood cells (WBCs), with other leukocytes or with body cells. The MHC determines donor compatibility for organ transplant, as well as one's susceptibility to autoimmune diseases.
In a cell, protein molecules of the host's own phenotype or of other biologic entities are continually synthesized and degraded. Each MHC molecule on the cell surface displays a small peptide (a molecular fraction of a protein) called an epitope. The presented self-antigens prevent an organism's immune system from targeting its own cells. The presentation of pathogen-derived proteins results in the elimination of the infected cell by the immune system.9

Synopsis
Swaminathan et al investigated the impact of changes in Omicron on a uniquely immunodominant spike-encoded CD8+ T cell epitope. They recruited 59 participants who had recovered from the first wave of COVID-19 in Australia in 2020 and, using SARS-CoV-2 encoded antigens for spike (S-1 pool) and ORF3A in HLA-A*29:02+ participants, analysed the pool of CD8+ T cell responses against native epitope YFPLQSYGF and Omicron specific epitope YFPLRSYSF.
They then assessed the immune response of vaccinated individuals from a pool of 104 participants also against native YFPLQSYGF and then Omicron YFPLRSYSF.
To investigate the impact of amino acid changes on another spike-encoded epitope, they also assessed T cell responses to the HLA-B*07:02-restricted epitope SPRRARSVA in five HLA-B*07:02-positive vaccine recipients.
APC-HLA-A*24:02/SARS-CoV-2 S (YFPLQSYGF) MHC Tetramer is a MHC tetramer of biotinylated peptide/MHC complex composed S-derived peptide of YFPLQSYGF sequence covering 489-497 and HLA-A*24:02 molecule. The pMHC tetramer recognizes CD8 T cells and can be used in the MHC tetramer analysis of individual antigen-specific T cells.10
In other words they combined the spike protein epitopes with MHC molecules and assessed the degrees of CD8+ T cell activation that occurred in response.
Abstract
The emergence of the SARS-CoV-2 Omicron variant has raised concerns of escape from vaccine-induced immunity. A number of studies have demonstrated a reduction in antibody-mediated neutralization of the Omicron variant in vaccinated individuals. Preliminary observations have suggested that T cells are less likely to be affected by changes in Omicron. However, the complexity of human leukocyte antigen genetics and its impact upon immunodominant T cell epitope selection suggests that the maintenance of T cell immunity may not be universal. In this study, we describe the impact that changes in Omicron BA.1, BA.2 and BA.3 have on recognition by spike-specific T cells. These T cells constitute the immunodominant CD8+ T cell response in HLA-A*29:02+ COVID-19 convalescent and vaccinated individuals; however, they fail to recognize the Omicron-encoded sequence. These observations demonstrate that in addition to evasion of antibody-mediated immunity, changes in Omicron variants can also lead to evasion of recognition by immunodominant T cell responses.
A conserved epitope is an epitope retained by multiple strains of a virus as the key target of a broadly neutralizing antibody.11
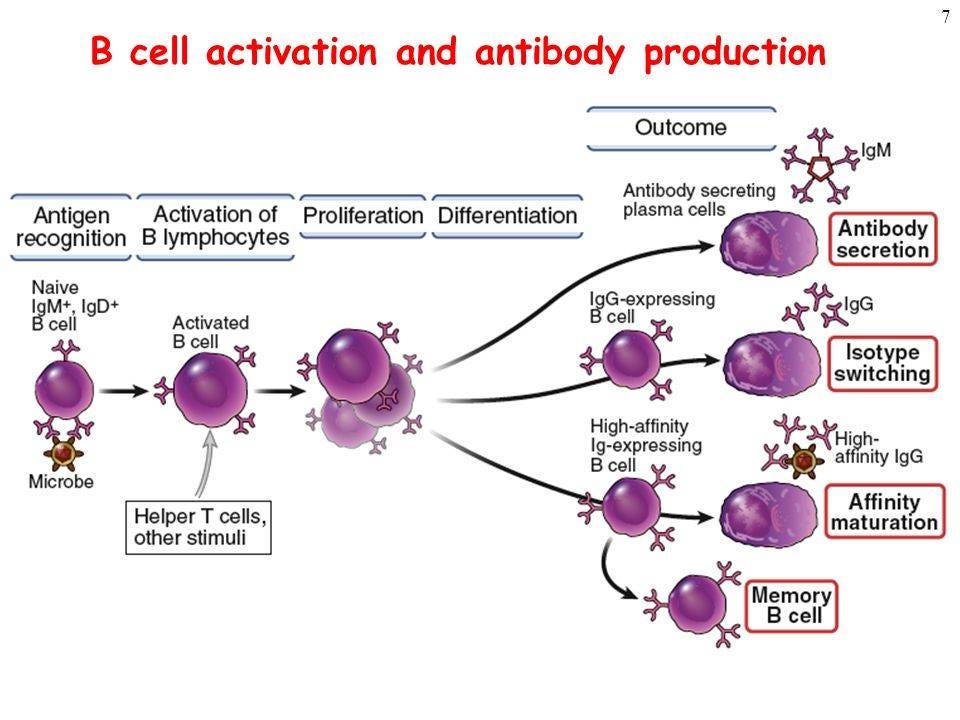
An effective immune response requires reactive T-cells12, not just antibodies from memory B cells which may or may not bind and neutralise the virus prior to being digested by phagocytes:13

Introduction
T cell immunity augments vaccine-mediated protection against SARS-CoV-2, particularly in the face of emerging variants such as Omicron and is likely to be critically important in individuals who fail to generate efficient neutralizing antibody responses due to underlying disease. Following the emergence of Omicron, studies have demonstrated that most T cell epitopes are conserved and the majority of vaccine recipients maintain T cell immunity. Nevertheless, evidence in convalescent individuals has demonstrated the capacity for variant strains to evade T cell responses. In this study, we sought to investigate the impact of changes in Omicron on a uniquely immunodominant spike-encoded CD8+ T cell epitope.
ORF3a (previously known as X1 or U274) is a gene found in coronaviruses of the subgenus Sarbecovirus, including SARS-CoV and SARS-CoV-2. It encodes an accessory protein about 275 amino acid residues long, which is thought to function as a viroporin. It is the largest accessory protein and was the first of the SARS-CoV accessory proteins to be described.14
A cytotoxic T cell (also known as TC, cytotoxic T lymphocyte, CTL, T-killer cell, cytolytic T cell, CD8+ T-cell or killer T cell) is a T lymphocyte (a type of white blood cell) that kills cancer cells, cells that are infected by intracellular pathogens (such as viruses or bacteria), or cells that are damaged in other ways.
Most cytotoxic T cells express T-cell receptors (TCRs) that can recognize a specific antigen. An antigen is a molecule capable of stimulating an immune response and is often produced by cancer cells, viruses, bacteria or intracellular signals. Antigens inside a cell are bound to class I MHC molecules, and brought to the surface of the cell by the class I MHC molecule, where they can be recognized by the T cell. If the TCR is specific for that antigen, it binds to the complex of the class I MHC molecule and the antigen, and the T cell destroys the cell.
In order for the TCR to bind to the class I MHC molecule, the former must be accompanied by a glycoprotein called CD8, which binds to the constant portion of the class I MHC molecule. Therefore, these T cells are called CD8+ T cells.
The affinity between CD8 and the MHC molecule keeps the TC cell and the target cell bound closely together during antigen-specific activation. CD8+ T cells are recognized as TC cells once they become activated and are generally classified as having a pre-defined cytotoxic role within the immune system. However, CD8+ T cells also have the ability to make some cytokines, such as TNF-α and IFN-γ, with antitumour and antimicrobial effects.15

The T helper cells (Th cells), also known as CD4+ cells or CD4-positive cells, are a type of T cell that play an important role in the adaptive immune system. They aid the activity of other immune cells by releasing cytokines. They are considered essential in B cell antibody class switching, breaking cross-tolerance in dendritic cells, in the activation and growth of cytotoxic T cells, and in maximizing bactericidal activity of phagocytes such as macrophages and neutrophils. CD4+ cells are mature Th cells that express the surface protein CD4.
Unvaccinated human leukocyte antigen HLA-A*29:02+ participants generated significant CD8+ T cell responses to native epitope YFPLQSYGF, but a very poor response to omicron epitope YFPLRSYSF.

However, conserved epitope ORF3a mediated the immunodominant response in matrix pools 7, 8 and 15, so some individuals still mounted an active CD8+ T cell response despite the mutations in Omicron:

IFN-γ, or type II interferon, is a cytokine that is critical for innate and adaptive immunity against viral, some bacterial and protozoan infections. IFN-γ is an important activator of macrophages and inducer of major histocompatibility complex class II molecule expression. Aberrant IFN-γ expression is associated with a number of autoinflammatory and autoimmune diseases. The importance of IFN-γ in the immune system stems in part from its ability to inhibit viral replication directly, and most importantly from its immunostimulatory and immunomodulatory effects. IFN-γ is produced predominantly by natural killer cells (NK) and natural killer T cells (NKT) as part of the innate immune response, and by CD4 Th1 and CD8 cytotoxic T lymphocyte (CTL) effector T cells once antigen-specific immunity develops as part of the adaptive immune response. IFN-γ is also produced by non-cytotoxic innate lymphoid cells (ILC), a family of immune cells first discovered in the early 2010s.16
Note the response to viral N protein in unvaccinated participants. We know from other studies that naive vaccinated cohorts are unable to generate any T cell response from exposure to highly conserved nucleocapsid protein N protein unless previously infected:
“Next, we examined cellular immunity and therefore carried out interferon-γ release assays (IGRA) utilising peptide pools that mapped the spike (S) and the nucleocapsid (N) proteins, respectively. As the S protein, which contains the receptor binding domain, is the vaccine target of BNT162b2, co-incubation of lithium heparin blood with spike peptide pools (pepS) resulted in marked IFN-γ responses in all test subjects. Notably, vaccine recipients with a prior COVID-19 history showed significantly stronger IGRA reactivities (1117•0 [95% CI 836•5–1491•0] pg/mL) indicating the presence of more SARS-CoV-2 spike protein-specific T cells compared to COVID-19 naïve vaccine recipients (620•6 [95% CI 460•2–836•9] pg/mL, t-test p<0•05; Fig. 2a). After the second vaccination, all test subjects demonstrated IFN-γ release upon pepS stimulation, whereas co-incubation with the nucleocapsid peptide pools (pepN) resulted almost exclusively in an IFN-γ release in subjects that had recovered from COVID-19 (1•1 [95% CI 0•3–4•0] pg/mL vs. 732•3 [95% CI 450•0–1192•0) pg/mL], t-test p<0•0001; Fig. 2a)”17

Curiously, the authors of our review paper don’t provide this type of heatmap for vaccinated individuals, but their data does show it is dominated by anti-S CD8+ T cells that are non-neutralising to omicron mutations, otherwise known as original antigenic sin.

Results and discussion
Our group recruited 59 COVID-19 convalescent participants during the initial 2020 wave of the pandemic in Australia. All participants were typed for human leukocyte antigens (HLA, Supplementary Table 1) and SARS-CoV-2-specific T cells were expanded from peripheral blood mononuclear cells (PBMC) using SARS-CoV-2-encoded antigens. In this analysis, we noted striking immunodominant CD8+ T cell responses to both spike (S-1 pool) and ORF3A in HLA-A*29:02+ participants (Fig. 1A, Supplementary Fig. 1). We detected a median S-1 pool-specific CD8+ T cell response of 44.6% in HLA-A*29:02+ participants compared to 0.85% in HLA-A*29:02− participants (Fig. 1B). The ORF3A-specific CD8+ T cell response was detected at a median of 31.2% in HLA-A*29:02+ participants compared to 1.47% in HLA-A*29:02- participants. Using an overlapping peptide matrix, we identified that the spike-specific T cell response targeted spike matrix pools 5, 6 and 23 (Fig. 1C), corresponding to overlapping peptides GFNCYFPLQSYGFQP and YFPLQSYGFQPTNGV (Supplementary Fig. 2A). Peptide minimization and HLA restriction (Supplementary Fig. 2B) showed the immunodominant response to be encoded by a HLA-A*29:02-restricted epitope, YFPLQSYGF (Fig. 1D). The immunodominant response in matrix pools 7, 8 and 15 from ORF3a (Fig. 1E) corresponded to overlapping peptides VVLHSYFTSDYYQLY and SYFTSDYYQLYSTQL (Supplementary Fig. 2C), which contain the previously published HLA-A*29:02-restricted epitope YFTSDYYQLY (Fig. 1F)12.

SARS-CoV-2 specific T cells from COVID-19 convalescent participants were expanded from PBMC using SARS-CoV-2 encoded antigens. PBMCs were cultured for 2 weeks in the presence of IL-2 and assessed for IFN-g responses following recall with the specific antigen. A Heatmap of the magnitude of SARS-CoV-2 antigen-specific T cell responses in COVID-19-convalescent individuals (HLA-A*29:02+, n = 4; HLA-A*29:02-, n = 55). B Frequency of S-1- and ORF3A-specific IFN-γ-producing CD8+ T cells in HLA-A*29:02+ and HLA-A*29:02– participants. Whiskers represent minimum to maximum values, boxes represent the 25th to 75th percentile, line represents the median. C Frequency of IFN-γ-producing CD8+ T cells in response to S-1 peptide matrix pools in HLA-A*29:02+ participants (n = 2). D Frequency of IFN-γ-producing CD8+ T cells in response to the YFPLQSYGF epitope in HLA-A*29:02+ participants. E Frequency of IFN-γ-producing CD8+ T cells in response to the ORF3A peptide matrix pools in HLA-A*29:02+ participants (n = 2). F Frequency of IFN-γ-producing CD8+ T cells in response to the YFTSDYYQLY epitope in HLA-A*29:02+ participants. G Prevalent HLA-A*29:02 epitope sequences in SARS-CoV-2 variants of concern. H Frequency of IFN-γ-producing CD8+ T cells in response to serial dilutions of YFPLQSYGF and YFPLRSYSF in HLA-A*29:02+ participants. Data represent mean and standard deviation of duplicates. Source data are provided as a source data file.
Next, they generated T cells specific for conserved, cognate (ie recognised) epitope YFPLQSYGF from three unvaccinated COVID-19-convalescent participants and performed intracellular cytokine analysis using 10-fold serial peptide titration with both the cognate epitope and Omicron variant, but this time assessing for single point mutated peptides, and again demonstrated poor reactivity against both:
Analysis of the amino acid sequences of the HLA-A*29:02 epitopes in SARS-CoV-2 variants showed that while the ORF3A-encoded epitope was conserved, the Omicron BA.1 variant contained amino acid substitutions at positions 5 and 8 in the spike-encoded epitope (Fig. 1G). The substitution at position 5 was also evident in the Omicron BA.2 and BA.3 variants, whilst the Omicron BA.4, BA.5 and BA.2.75 variants had reverted to the wild-type sequence. Although not anchor residues for A*29:02, the substitutions, particularly the non-conserved glutamine (Q) to arginine (R) change at position 5, have the potential to significantly impact T cell receptor (TCR) engagement. To address this, we generated T cells specific for the YFPLQSYGF epitope from three COVID-19-convalescent participants and performed intracellular cytokine analysis using 10-fold serial peptide titration with both the cognate epitope and Omicron variant. Despite strong functional avidity directed towards the cognate peptide (Supplementary Fig. 3A), we saw poor reactivity against the Omicron variant (Fig. 1H, Supplementary Fig. 3B). To assess the impact of single amino acid substitution at position 5, YFPLRSYGF and position 8, YFPLQSYSF, functional avidity assays with the single mutant peptides were performed. We saw poor reactivity against the both YFPLRSYGF and YFPLQSYSF single mutated peptide in Q031 (Supplementary Fig. 3C) and Q043 (Supplementary Fig. 3D).
They then repeated the above analysis, but this time from a cohort of 104 participants, recruited following vaccination with either an adenovirus vector (Vaxzevria) or an mRNA (Cominarty, BNT162b2) vaccine.
The participant numbers are in single figures, but even so the dominant response is to the native epitope YFPLQSYGF, with a negligible response to omicron mutated epitope YFPLRSYSF.
We next assessed if individuals vaccinated against SARS-CoV-2 would show a similar pattern in their response to YFPLQSYGF. From a cohort of 104 participants, recruited following vaccination with either an adenovirus vector (Vaxzevria) or an mRNA (Cominarty) vaccine, we identified three participants with HLA-A*29:02 (Supplementary Table 2). PBMC from these individuals were isolated 28 days after their second vaccine dose, then stimulated with a spike peptide pool, the YFPLQSYGF epitope or the Omicron variant. We assessed expression of IFN-γ and TNF by CD8+ T cells using intracellular cytokine analysis to identify low-frequency T cell responses. Controls were unstimulated PBMC (no peptide) and PBMC incubated with a cell stimulation cocktail (CSC). Although low in abundance, IFN-γ+TNF+ CD8+ T cell responses against the spike overlapping peptide pools (0.006%, 0.009%, and 0.002% above no peptide) and the YFPLQSYGF epitope (0.046%, 0.012%, and 0.006% above no peptide) were detected in PBMC from all three individuals (Fig. 2A). Similar observations were evident following analysis of IFN-γ alone (Supplementary Fig. 4A). Conversely, CD8+ T cells capable of recognizing the Omicron variant were not detected. To validate these observations, we generated T cells from these three participants by stimulating with the spike peptide pools and culturing in the presence of IL-2 for 14 days. In addition, we set up cultures from a fourth individual in our vaccine cohort who is HLA-A*29:11+. Spike-specific CD8+ T cell responses were observed in all four T cell expansions (Fig. 2B, Supplementary Fig. 4B). All four cultures were dominated by T cells specific for YFPLQSYGF, but displayed no recognition of the Omicron variant. In the three HLA-A*29:02+ cultures, the frequency of the YFPLQSYGF-specific CD8+IFN-γ+TNF+ response was comparable to the response against whole spike protein, demonstrating similar immunodominance to that seen in convalescent individuals. To generate variant-specific T cells, we stimulated PBMC with either YFPLRSYSF or the Omicron variant and assessed functional avidity after 14 days in culture. While the Omicron epitope failed to induce the expansion of T cells in these donors, YFPLRSYSF-specific T cells were detected in cultures from all four vaccine recipients (Fig. 2C, D). To confirm the immunodominance of the YFPLQSYGF response in HLA-A*29 individuals, we compared the response to 16 other HLA-A*29:02-restricted epitopes identified from the immune epitope database and prediction resource (IEDB) (Supplementary table 3). While YFPLQSYGF T cell responses were detected in all four volunteers assessed, we could only detect low-frequency subdominant responses to SANNCTFEY (2.83%) and CVADYSVLY (0.66%) in GR049, and LTDMIAQY (0.84%) in GR166, confirming the immunodominance of the YFPLQSYGF in HLA-A29*02-positive individuals (Fig. 2E).
Recognition of the immunodominant HLA-A*29:02 in vaccinated participants with omicron YFPLRSYSF is in blue, as measured by IFN-γ and TNFa producing CD8+ T cells.
As commented earlier a lost opportunity here was the failure to provide a heatmap of responses by vaccinated participants to additional viral epitopes such as ORF or N-protein:

A Flow cytometry plots displaying the frequency of IFN-γ-producing CD8+ T cells from PBMC of vaccinated HLAA*29:02+ individuals. B Frequency of IFN-γ-producing CD8+ T cells following in vitro culture of PBMC from four vaccinated individuals (A*29:02 n = 3; A*29:11 n = 1) with spike overlapping peptide pools. C Frequency of IFN-γ-producing CD8+ T cells following in vitro stimulation with either YFPLQSYGF or YFPLRSYSF and recall with cognate peptide. D Frequency of IFN-γ-producing CD8+ T cells in response to serial dilutions of YFPLQSYGF. Data represents mean and standard deviation of duplicates. E Heatmap of frequency of IFN-γ-producing CD8+ T cells of spike peptide pool, YFPLQSYGF and published HLA-A29*02 from four vaccinated individuals (A*29:02 n = 3; A*29:11 n = 1). F Crystal structure of the HLA-A*29:02-YFPLRSYSF complex with the HLA heavy chain in white cartoon and the peptide in red cartoon and stick. The P8-Gly Cα atom is represented by a sphere, and the hydrogen bond by a red dashed line. G Model of the YFP variant peptide (blue cartoon and stick) based on the YFP peptide conformation showing the P5-Arg and P8-Ser substitutions in yellow stick. The HLA Met97, Tyr99, and Arg114 are represented as stick and transparent surface to show the steric clashes with the P5-Arg occurring if the variant adopted the same conformation as the cognate peptide. Source data are provided as a source data file.
Next, they investigated the impact of the Omicron mutation on peptide presentation by HLA-A*29:02. They refolded native YFPLQSYGF with the HLA, crystalised it and assessed its thermal stability, or how structurally stable the peptide is.
The omicron mutation at position 5 can have an impact on the conformation of the peptide, destabilize the peptide human leukocyte antigen (pHLA) complex and weaken its binding to HLA-A*29:02, thus suppressing any potential CD8+ T cell response:
Given the differences in the T cell response between YFPLQSYGF and its variant, we wanted to understand the impact of the Omicron mutation on peptide presentation by HLA-A*29:02. YFPLQSYGF peptide was refolded with HLA-A*29:02 and then crystallized to understand the basis of peptide presentation (Supplementary Table 4). The HLA-A*29:02 molecule adopted the canonical fold of HLA molecules with YFPLQSYGF (Supplementary Fig. 5A) and in line with the thermal stability observed for the YFP-HLA-A*29:02 complex (Supplementary Table 5).The primary anchors of the YFPLQSYGF peptide at P2 and P9 are both phenylalanine residues that are favored for HLA-A*29:02-peptide binding motif from netMHCpan4.1 website (Fig. 2F). The motif extracted from the netMHCpan4.1 website reports the naturally presented peptide are those which are derived from eluted mass spectrometry data (Supplementary Fig. 6A) however, MHC binder peptides are derived from the binding affinity data from in vitro expansion (Supplementary Fig. 6B). The two residues of the peptide that are mutated in Omicron are at positions 5 and 8. While the substitution at P8 from a glycine to a serine is unlikely to impact peptide presentation and conformation (Fig. 2G), the mutation at position 5 can have an impact on the conformation of the peptide. The P5-Gln is buried in the cleft of HLA-A*29:02, and modeling of a P5-Arg shows a steric clash of the large and charged side chain (Fig. 2G) with residues in the binding cleft due to the presence of large amino acid residues Met97, Tyr99 and Arg114 that occupy the β-sheet floor of HLA-A*29:02. The P5 mutation in Omicron is likely to destabilize the peptide human leukocyte antigen (pHLA) complex and could also weaken its binding to HLA-A*29:02.
To confirm this, they refolded the HLA-A*29:02 molecule, but this time with the YFP mutant derived from Omicron. They then analysed the 2 single point mutations individually and again performed stability assays which appeared to confirm that the mutation of the P5Q by P5R is destabilizing the pHLA complex:
To confirm this hypothesis, we refolded the YFP mutant derived from Omicron with the HLA-A*29:02 molecule and performed a stability assay. The result shows a dramatic decrease of the pHLA complex stability by 14 °C due to the mutations in the Omicron-derived peptide (Supplementary Table 5). This confirmed that the binding of the peptide to HLA-A*29:02 is reduced due to the mutations occurring in the Omicron variant. It is likely that the mutation of the secondary anchor residue P5-Gln is responsible for the decreased Tm value observed, as the P8-Gly is solvent exposed and would not stabilize the peptide but could impact directly on T cell binding. To confirm this hypothesis we have produced single mutant of the YFP peptide at positions 5 and 8 to test the impact of each mutation. Each mutated peptides were able to be refolded with the HLA-A*29:02 molecule, and their stability was determined. The single mutant YFP-P8S in complex with HLA-A*29:02 exhibited a stability close to the one observed for the YFP peptide, decreased by 4 °C (Supplementary Table 5 and Supplementary Fig. 7). Interestingly, both the single mutant YFP-P5R and the omicron variant of YFP both exhibit a similar Tm of ~46–47 °C which is 15 °C lower than the stability of HLA-A*29:02-YFP complex. This shows that indeed the mutation of the P5Q by P5R is destabilizing the pHLA complex.
Next, they investigated the impact of amino acid changes on another spike-encoded epitope, SPRRARSVA, on the T cell activation of five HLA-B*07:02-positive vaccine recipients and also against other HLA-A*02:01–restricted epitopes:
To investigate the impact of amino acid changes on another spike-encoded epitope, we assessed T cell responses to the HLA-B*07:02-restricted epitope SPRRARSVA in five HLA-B*07:02-positive vaccine recipients (Supplementary Table 2). This epitope contains mutations detectable in multiple variants that alter the HLA-B*07:02 anchor residue at P2 (Supplementary Table 6). While these changes ablated the ability of T cells from vaccinated individuals to recognize the variant peptides, other spike-specific T cell responses were detected, including those against the conserved HLA-A*02:01–restricted epitope YLQPRTFLL (Supplementary Fig. 4C), demonstrating the complex nature of T cell immunodominance in HLA-distinct individuals.
Response to omicron YFPLRSYSF is in blue, if at all:
To conclude, the authors then consider the risk of original antigenic sin for vaccinated individuals and the effects of population levels of HLA-A29 on mounting a significant CD8+ T cell response.
They do suggest repeating the investigations with a much larger cohort:
Despite the potentially large number of antigenic targets in the spike protein, our observations suggest that the immunodominance profile in HLA-A29-positive individuals is associated with a skewed response towards a single dominant epitope. While it remains to be determined if this could impact susceptibility in vaccinated individuals, and the current study was limited by the number of individuals assessed, it was clear that changes in the amino acid sequence ablated T cell activation of this immunodominant response. Although HLA-A29 was present in only 6.8% and 3.8% of our convalescent and vaccinated cohort, respectively, HLA-A29 frequencies have been reported as high as 24% in some populations in Africa (allelefrequencies.net, Supplementary Table 7). However, knowledge on the impact of HLA-types on outcome of SARS-CoV-2 infection is currently limited, and the impact of changes in the omicron variants to risk in HLA-A*29 positive individuals requires investigation in a much larger cohort. Nevertheless, these analyses do highlight the impact genetic variation in newly emerging SARS-CoV-2 variants can have upon recognition by immunodominant T-cell responses.
Experimental methods are then detailed, which I will exclude for brevity.
Acknowledgements
This work was supported by generous donations to the QIMR Berghofer COVID-19 appeal and the Medical Research Future Fund (MRFF, APP2005654). SS is supported by Australian Government Research Training Program Scholarship and QIMR Berghofer Top-Up Scholarship award. SG is an NHMRC SRF-A Fellow (1159272). KRS is supported by an NHMRC Investigator Grant (2007919). We would like to thank all of the participants who generously donated their blood for this study. This research was undertaken in part using the MX2 beamline at the Australian Synchrotron, part of ANSTO, and made use of the Australian Cancer Research Foundation (ACRF) detector.
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
Closing remarks
An opportunity was lost in not conducting a similar assessment for ablation of CD8+ T cell recognition in vaccinated vs unvaccinated cohorts against the Delta variant in 2021.
Original antigenic sin is difficult to reverse if each subsequent exposure to either live viral variants or a booster stimulates a backboost of immunodominant but non-neutralising T-cells, or no response at all.
This could have been avoided in the population at large and instead broad spectrum anti-viral therapeutics been administered to those at particular risk due to immunosenescence.18
Another factor the authors do not consider is T cell exhaustion or anergy19 due to repeated overstimulation with vaccinal spike protein. This can be a co-factor to OAS, further multiplying the risk to affected individuals. From Molecular regulation of T-cell anergy by Zheng et al (2008):
Abstract
The activation of T cells is tightly controlled by many positive and negative regulatory processes. This fine-tuning allows productive immunity to pathogens while minimizing the risk of autoimmunity. One negative regulatory mechanism is clonal anergy, which is a hyporesponsive state that occurs when T cells are activated through the T-cell antigen receptor in the absence of appropriate co-stimulatory signals. Recent studies have confirmed a crucial role for defective Ras activation in mediating this hyporesponsive state. Diminished Ras activation can, in part, be explained by the upregulated expression of diacylglycerol kinases (DGKs), which phosphorylate diacylglycerol and restrict Ras guanyl releasing protein 1 (RasGRP1)-dependent activation of Ras. Increased expression of DGKs is probably transcriptional and is accompanied by augmented expression of additional negative regulators, including the transcription factors early growth response (Egr) 2 and Egr3, and the E3 ubiquitin ligases known as gene related to anergy in lymphocytes (GRAIL) and Casitas B-cell lymphoma-b (Cbl-b). A model is emerging for how these factors are regulated to control T-cell responsiveness.
Keywords: T-cell activation, signal transduction, Ras, diacylglycerol kinase, anergy
…They found that when T cells encountered antigens presented by chemically fixed APCs—which presumably failed to upregulate co-stimulatory ligands—the T cells proliferated poorly and made less interleukin-2 (IL-2) than when live APCs were used. In addition, when the T cells were recovered and restimulated, they were found to be hyporesponsive. The induction of anergy has also been observed with other agents that engage the TCR complex alone, such as purified MHC–peptide complexes displayed on a planar lipid bilayer, anti-CD3 monoclonal antibody, the mitogen concanavalin A, or peptides presented by inefficient APCs such as small resting B cells (Gajewski et al, 1994a). In addition to a nearly complete block in IL-2 production and proliferation, anergic T cells show reduced secretion of other cytokines such as interferon-γ (IFN-γ) and IL-3. The anergic state can be long-lived and stable for weeks, but in these model systems it is reversible by the addition of exogenous cytokines such as IL-2 or IL-15 (Beverly et al, 1992; Essery et al, 1988). This reversibility has motivated further investigation of anergy in the settings of cancer and chronic infection, in which restoration of correct T-cell function is one of the main therapeutic goals.
An anergy-like state can also be induced in vitro by altered peptide ligands, which are peptides with minor sequence differences from native agonist antigenic peptides and that give lower avidity TCR ligation (Sloan-Lancaster et al, 1993). Engagement of the TCR with altered peptide ligands even in the presence of co-stimulation induces a state of hyporesponsiveness that resembles anergy. Recently, Korb and colleagues reported that low doses (fewer than 10 peptide/MHC complexes per APC) of agonist peptides can also induce an anergy-like T-cell dysfunctional state (Korb et al, 1999; Mirshahidi et al, 2001). It is not clear whether the molecular regulation of these types of T-cell dysfunction is the same as that which has been determined for classical anergy—that is, agonist TCR ligation in the absence of co-stimulation.
From Innate immune suppression by SARS-CoV-2 mRNA vaccinations: The role of G-quadruplexes, exosomes, and MicroRNAs by Seneff et al (2022):20
Bearing in mind the multiple mutations that SARS-CoV-2 develops, as for example in the Brazil outbreaks (Timmers et al., 2021), an effective immune response that prevents the spread of SARS-CoV2 mutants necessarily involves the development of a robust IFN-I response as a part of the innate immune system. This response also requires the involvement of a functional NF-κB response. Unfortunately, spike glycoprotein overexpression dismantles the NF-κB pathway responses, and this molecular event can be augmented by spike-protein-coding mRNAs (Kyriakopoulos and McCullough, 2021; Jiang and Mei, 2021).
The failure of current omicron focused bivalent boosters due to OAS is a strong indicator that current public health policy needs to pivot instead to safe and effective, broad spectrum antivirals with accompanying immune supportive diets and supplementation as a matter of priority. Ivermectin would provide a good foundation for this, along with other therapeutics reviewed separately in this Substack channel.
Preprint by Wang et al (2022), Antibody responses to Omicron BA.4/BA.5 bivalent mRNA vaccine booster shot:21
Abstract
The SARS-CoV-2 Omicron variant and its numerous sub-lineages have exhibited a striking ability to evade humoral immune responses induced by prior vaccination or infection. The Food and Drug Administration (FDA) has recently granted Emergency Use Authorizations (EUAs) to new bivalent formulations of the original Moderna and Pfizer mRNA SARS-CoV-2 vaccines that target both the ancestral strain as well as the Omicron BA.4/BA.5 variant. Despite their widespread use as a vaccine boost, little is known about the antibody responses induced in humans. Here, we collected sera from several clinical cohorts: individuals after three or four doses of the original monovalent mRNA vaccines, individuals receiving the new bivalent vaccines as a fourth dose, and individuals with BA.4/BA.5 breakthrough infection following mRNA vaccination. Using pseudovirus neutralization assays, these sera were tested for neutralization against an ancestral SARS-CoV-2 strain, several Omicron sub-lineages, and several related sarbecoviruses. At ~3-5 weeks post booster shot, individuals who received a fourth vaccine dose with a bivalent mRNA vaccine targeting BA.4/BA.5 had similar neutralizing antibody titers as those receiving a fourth monovalent mRNA vaccine against all SARS-CoV-2 variants tested, including BA.4/BA.5. Those who received a fourth monovalent vaccine dose had a slightly higher neutralizing antibody titers than those who received the bivalent vaccine against three related sarbecoviruses: SARS-CoV, GD-Pangolin, and WIV1. When given as a fourth dose, a bivalent mRNA vaccine targeting Omicron BA.4/BA.5 and an ancestral SARS-CoV-2 strain did not induce superior neutralizing antibody responses in humans, at the time period tested, compared to the original monovalent vaccine formulation.
Boosting with a new bivalent mRNA vaccine targeting both BA.4/BA.5 and an ancestral SARS-CoV-2 strain did not elicit a discernibly superior virus-neutralizing antibody responses compared boosting with an original monovalent vaccine. These findings may be indicative of immunological imprinting, although follow-up studies are needed to determine if the antibody responses will deviate in time, including the impact of a second bivalent booster.
No advantage is conferred by including BA.4/BA.5 specific spike protein epitopes in a bivalent booster.
A note about the high ID50’s against D614G:
The D614G mutation in the viral spike protein is one of the earliest significant mutations, occurring in late January to early February 2020, and viruses containing glycine residue at position 614 became the dominant form of the virus globally, replacing the initial strain identified in China, by June 2020.22
However, there is evidence suggesting that the possibility of D614G mutation affecting the vaccine efficacy is very low. Because the mutation is not in the receptor-binding domain of the spike protein, it is less likely to affect the ability of the domain to induce host immune responses, which is believed to be a prerequisite for antibody-mediated neutralization of the virus.
Moreover, a large portion of ongoing vaccines are developed against the receptor-binding domain, and thus, D614G mutation is not supposed to have any effect on vaccine efficacy.
Another important observation is that convalescent sera infected with D614 containing viruses have been found to neutralize viruses containing G614, and vice versa. This indicates that D614G mutation does not alter antibody-mediate immune responses.
The Y-axis antibody neutralisation ID50 titers are to a log scale, so each gradation denotes a tenfold increase (or decrease) in measured serum neutralisation.
Antibodies do not equal immunity, and this isn’t a measure of T-cell, ORF3A or nucleocapsid protein responses, but it is useful for comparative purposes:

(A) Neutralization ID50 titers of serum samples from “3 shots WT”, “BA.4/BA.5 breakthrough”, “4 shots WT”, and “3 shots WT + bivalent” cohorts. Values above the symbols denote the geometric mean ID50 titers, and values on the lower left indicate the sample size (n). The limit of detection is 100 (dotted line). Wild-type (WT) shots refer to monovalent mRNA vaccine doses. “3 shots WT,” sera from individuals vaccinated with three doses of the WT mRNA vaccine; “BA.4/BA.5 breakthrough,” sera from individuals with BA.4 or BA.5 infection following WT mRNA vaccination; “4 shots WT,” sera from individuals vaccinated with four doses of the WT mRNA vaccine; “3 shots WT + bivalent,” sera from individuals vaccinated with three doses of the WT mRNA vaccine and subsequently one dose of a BA.4/BA.5 bivalent mRNA vaccine. (B) Comparison of antibody responses induced by a fourth dose of the original WT mRNA vaccine versus a fourth dose of a BA.4/BA.5 bivalent mRNA vaccine. Comparisons were made by Mann-Whitney tests. *p < 0.05; **p < 0.01; ***p < 0.001. Values above the symbols denote the geometric mean ID50 titers.
From the presentation Pfizer/BioNTech COVID-19 Omicron-Modified Vaccine Options. Note use of the log scale, the inconsistency in the spread of titers (hilighted) and that all 10 Moderna boosted mice got infected when challenged with Ba.5 variant virus.
Titers in the 8 bivalent booster treated mice (purple bar on far right) ranged from 300 to 22,000:23
References
Swaminathan, S., Lineburg, K.E., Panikkar, A. et al. Ablation of CD8+ T cell recognition of an immunodominant epitope in SARS-CoV-2 Omicron variants BA.1, BA.2 and BA.3. Nat Commun 13, 6387 (2022). https://doi.org/10.1038/s41467-022-34180-1
https://www.nature.com/articles/s41467-022-34180-1#rightslink
Pan K (2011) Understanding Original Antigenic Sin in Influenza with a Dynamical System. PLoS ONE 6(8): e23910. https://doi.org/10.1371/journal.pone.0023910
Cell ablation - Wikipedia
Leukocyte | definition of leukocyte by Medical dictionary (thefreedictionary.com)
Epitope - Wikipedia
Brézin AP, Monnet D, Cohen JH, Levinson RD. HLA-A29 and birdshot chorioretinopathy. Ocul Immunol Inflamm. 2011 Dec;19(6):397-400. doi: 10.3109/09273948.2011.619295. PMID: 22106906.
HLA-A29 - Wikipedia
Human leukocyte antigen - Wikipedia
Major histocompatibility complex - Wikipedia
https://en.wikipedia.org/wiki/Major_histocompatibility_complex
APC-HLA-A*24:02/SARS-CoV-2 S (YFPLQSYGF) MHC Tetramer - Creative Biolabs
https://www.creativebiolabs.net/apc-hla-a-24-02-sars-cov-2-s-489-497-mhc-tetramer-135066.htm
Ren J, Ellis J, Li J. Influenza A HA's conserved epitopes and broadly neutralizing antibodies: a prediction method. J Bioinform Comput Biol. 2014 Oct;12(5):1450023. doi: 10.1142/S0219720014500231. Epub 2014 Sep 10. PMID: 25208658.
T cells on the front line of COVID-19 response
https://www.news-medical.net/news/20200917/T-cells-on-the-front-line-of-COVID-19-response.aspx
Mechanism of antibody production
http://sciencemission.com/site/index.php?page=news&type=view&id=immunology/mechanism-of-antibody
Cytotoxic T cell - Wikipedia
Interferon gamma - Wikipedia
Zollner A, Watschinger C, Rössler A, Farcet MR, Penner A, Böhm V, Kiechl SJ, Stampfel G, Hintenberger R, Tilg H, Koch R, Antlanger M, Kreil TR, Kimpel J, Moschen AR. B and T cell response to SARS-CoV-2 vaccination in health care professionals with and without previous COVID-19. EBioMedicine. 2021 Aug;70:103539. doi: 10.1016/j.ebiom.2021.103539. Epub 2021 Aug 12. PMID: 34391087; PMCID: PMC8358275.
Immunosenescence - Wikipedia
Zheng Y, Zha Y, Gajewski TF. Molecular regulation of T-cell anergy. EMBO Rep. 2008 Jan;9(1):50-5. doi: 10.1038/sj.embor.7401138. PMID: 18174897; PMCID: PMC2246614.
Seneff S, Nigh G, Kyriakopoulos AM, McCullough PA. Innate immune suppression by SARS-CoV-2 mRNA vaccinations: The role of G-quadruplexes, exosomes, and MicroRNAs. Food Chem Toxicol. 2022 Jun;164:113008. doi: 10.1016/j.fct.2022.113008. Epub 2022 Apr 15. PMID: 35436552; PMCID: PMC9012513.
Qian Wang, Anthony Bowen, Riccardo Valdez, Carmen Gherasim, Aubree Gordon, Lihong Liu, David D. Ho. Antibody responses to Omicron BA.4/BA.5 bivalent mRNA vaccine booster shot. (2022). bioRxiv 2022.10.22.513349; doi: https://doi.org/10.1101/2022.10.22.513349
https://www.biorxiv.org/content/10.1101/2022.10.22.513349v1.full.pdf
D614G Mutation in SARS-CoV-2 Spike Protein
Pfizer/BioNTech COVID-19 Omicron-Modified Vaccine Options. (2022)











Fantastic job, you did a better job than I ever would, especially lately. Sharing it !
Edit: More and more evidence that some people will experience chronic infections like EBV, it is just a matter of time until they prove this 2020 point of mine (and many others, I doubt I was the only one making this point).
Thanks!
It should be called Original antigenic imprint OAI.