Self-amplifying mRNA (saRNA) Part 2, Episode V: The Pharma Empire Strikes Back
"In order to build your immunity, we need to destroy it"
This Substack is too long for email. Please click on the title to load in the app or a browser.
Reading time:
short movie - novelette - novella - novel - PhD thesis - War and Peace - U.S. Tax Code
Any extracts used in the following article are for non-commercial research and educational purposes only and may be subject to copyright from their respective owners.
Contents
Introduction
First off, apologies to Mr George Lucas.
[Recommended: sound on]
In Part 1 I translated and annotated Dr Stebel’s excellent walkthrough of the Japanese (pre-) registration documents for ARCT-154 (“Kostaive”). In short, many crucial safety studies were not conducted because associated pathologies were simply just ”… not expected to show”, or they had "… no particular special safety concerns".
If you want to know the pharmacokinetics of the drug, i.e. the movement of drug into, through, and out of the body, they cannot tell you in detail as no clinical studies were conducted.
Rabbit studies were done with the LNP carrier ATX-126, but that’s all we have, and these don’t translate directly to humans. It, or its expressed Spike protein, may accumulate in human brain meninges, the kidneys, heart, ovaries or testes and cause disease. But nobody will know until its too late to avoid harm.
Whilst reading around ARCT-154 I quickly discovered several features that raised red flags regarding cancer risk, as well as risks to new-borns. Many of these pathologies are shared with those of LNP-mRNA gene therapy agents.
The first thing that needs to be noted is that this Substack trilogy is NOT saying that saRNA technology can directly CAUSE cancer, as in initiate it like a Group 1 confirmed carcinogen1. There is a reason for this, in that the human genotoxicity research was also simply never done.
However, what these Substacks can do is review the current research literature, relate this to the inherent contradictions of the pro-tumour pathways that are being invoked and present the evidence that these products may act to promote cancer. This should be more than sufficient grounds for a full recall under the precautionary principle.
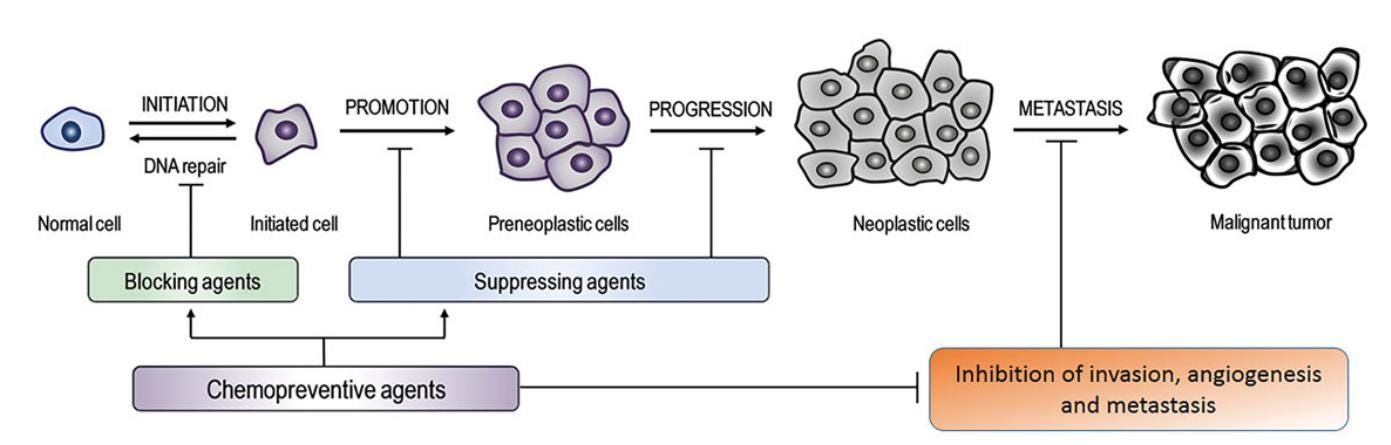
This should sound familiar to anyone who has read “Review: N1-methyl-pseudouridine (m1Ψ): Friend or foe of cancer?”2 as saRNA tech has the same requirement for the suppression of innate immunity in order to avoid premature degradation after detection by DNA and LNP sensing pattern recognition receptors (PRRs), the goal being to maximise production of the target protein.
This may be referred to as the “original sin” of the mRNA platform: in order to express enough antigens for long enough to generate an immune response it must, by necessity, suppress innate immune responses to itself. This design decision is not without consequences.
To keep the word count manageable this Substack will focus on two strategies that may be used by the saRNA platform to suppress innate immune signalling pathways and responses.
The first is by encoding for secondary proteins alongside the main immunogenic payload, and the second is by exploiting viral mutations which also demonstrate innate immunosuppression and increased protein expression.
Discussion
Innate inhibiting proteins (IIPs)
Why do viruses need to suppress the immune system of the host?
… For a virus to establish persistence in the host, it has to exploit the host immune system such that the active T-cell responses against the virus are curbed. On the other hand, the goal of the immune system is to clear the virus, following which the immune responses need to be downregulated, by a process known as immunoregulation. There are multiple known immunoregulatory mechanisms that appear to play a role in persistent viral infections.
… Most viruses that infect humans are usually cleared by the host immune system in a rapid (acute) manner, either (a) by the release of proinflammatory cytokines or (b) by killing the infected cells to limit the replication of the virus [1].
Such acute responses involve a first step innate immune response (macrophages, dendritic cells [DCs], and natural killer [NK] cells) [2], followed by the generation of adaptive immunity (T and B cells). Macrophages and DCs act as phagocytes thus eliminating the infected cells, and NK cells directly kill the infected cells.
In adaptive immunity, which kicks in 3–5 days after infection, presentation of viral antigen by macrophages and/or DCs or by infected cells that present viral peptides on host MHC molecules results in the activation of T-cells.
These T-cells then mount a viral antigen-specific immune response and kill the infected cells either in a cell–cell contact-dependent manner (perforin/granzyme mediated) or through the release of inflammatory cytokines TNF-α or IFN-γ.
These cytokines can also act on the virus directly and prevent its replication. Some of the viral antigen-specific T-cells then survive for a very long time creating a memory pool.
Most, if not all, viral infections trigger the production of interferon’s (IFNs) by the host immune system. IFNs then primarily limit viral replication to prevent damage to the infected cell.
Type-I IFNs are essential part of innate resistance to viral infections as is evident in the case B6 mice infected with MHV-1; absence of type I IFN-mediated signaling in IFN-αβR-KO mice results in progressive loss of body weight, culminating in the death of the mice by day 5 postinfection [3].
… Persistent viral infections, as would be expected, are a severe problem to humans, as is evident in the case infection with HIV; persistent infection with HIV leads to the exhaustion of CD4 T-cells thereby rendering the host susceptible to a variety of secondary infections, which ultimately lead to the demise of the host. However, such persistent infections, if are actively sought by the host immune system, due to the continued activation of T-cells can lead to severe immune pathology, wherein the host’s immune system destroys the host’s own tissue.
From: “Immunosuppressive Mechanisms During Viral Infectious Diseases“ (2010)
Immuno-suppressive mechanisms like these need to be exploited to help prevent primary vaccine failure and an intolerable number of adverse events, even allowing for regulators looking the other way.
“Improvement of In Vivo Expression of Genes Delivered by Self-Amplifying RNA Using Vaccinia Virus Immune Evasion Proteins” (2017) by Beissert et al3 describes the challenge.
Key takes:
(Emphasis in bold, as ever).
VACV: Vaccinia virus, from the pox family.
PKR: Protein kinase R, is an interferon-induced, double-stranded RNA-activated protein kinase that is activated by viral infections and inhibits viral protein synthesis.
IFNs: A group of signalling proteins that helps the body's immune system fight infection and other diseases, such as cancer.
E3, K3, and B18: Vaccinia virus immune evasion proteins.
eIF2α: Eukaryotic translational initiation factor 2. It is required for most forms of eukaryotic translation initiation, i.e. for protein synthesis.
I explain the function of these in part 3.
This study demonstrates that the expression of saRNA is substantially improved by co-transferring mRNA encoding VACV proteins inhibiting interferons and PKR.
By comparison of saRNA translation in BHK21 and HFF cells, it is demonstrated that the impact of PKR and IFN activation on protein translation is in the range of two orders of magnitude and thus substantial, even if accounting for the different growth kinetics and physiology of the two cell lines.
To reduce pattern recognition receptor stimulation and unleash suppressed saRNA translation, this study co-delivered non-replicating mRNA encoding vaccinia virus immune evasion proteins E3, K3, and B18.
It was shown that E3 is far superior to K3 or B18 as a highly potent blocker of PKR activation and of interferon (IFN)-β upregulation. B18, in contrast, is superior in controlling OAS1, a key IFN-inducible gene involved in viral RNA degradation.
By combining all three vaccinia proteins, the study achieved significant suppression of PKR and IFN pathway activation in vitro and enhanced expression of saRNA-encoded genes of interest both in vitro and in vivo.
Moreover, by complex formation, E3 affects PKR homodimerization,24 a prerequisite for its activation, and it was shown to inhibit OAS1 directly.25
K3 as an eIF2α mimic reduced the phosphorylation of eIF2α to some extent, but apparently not enough to unleash the full potential of saRNA translation.
B18 is a type I IFN binding protein and, as expected, it did not alter PKR and eIF2α phosphorylation. It was found that B18 very effectively neutralized co-secreted IFNs and thus eliminated interferon receptor signaling and induction of IFN responsive genes such as OAS1.
Synergistic effects by combining all three immunosuppressive viral proteins:
Even though the single-agent effects of K3 and B18 on reporter gene translation and protein production were marginal in vitro, as compared to E3, it was found that when all three were combined, K3 and B18 added an incremental improvement of saRNA translation. Therefore, the three VACV immune evasion proteins were combined for the subsequent in vivo setting.
In summary, this study demonstrates that mRNA based co-delivery of viral immune evasion proteins results in a log scale increase of saRNA translation. This RNA-based approach can ease the tasks of producing sufficient amounts of vaccines for large cohorts of patients, for example in the case of recurrent viral infections such as influenza or emerging pathogens with pandemic potential such as Ebola or Zika.
And in 2020, in “Innate Inhibiting Proteins Enhance Expression and Immunogenicity of Self-Amplifying RNA”, Blakney et al. conducted in vivo studies of rabbits, mice and rats to investigate how virus-associated immune-suppressing proteins could be used to enhance saRNA protein expression4.
A co-author for this paper was NewsGuard’s “Fact-Checker” consultant Robin J Shattock. His name comes up as one of the owners of an important saRNA patent concerning IIPs. This will be discussed further in the final part of the trilogy.

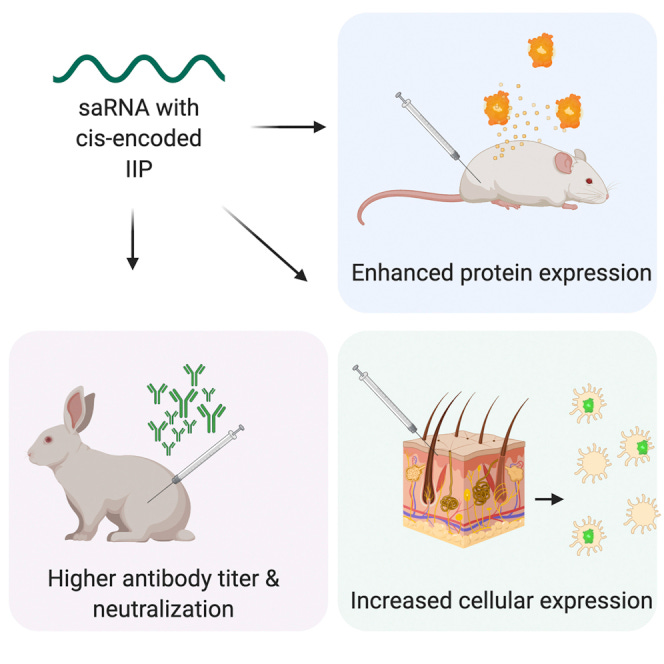
Self-amplifying RNA (saRNA) is a cutting-edge platform for both nucleic acid vaccines and therapeutics. saRNA is self-adjuvanting, as it activates types I and III interferon (IFN), which enhances the immunogenicity of RNA vaccines but can also lead to inhibition of translation.
In this study, we screened a library of saRNA constructs with cis-encoded innate inhibiting proteins (IIPs) and determined the effect on protein expression and immunogenicity.
PIV-5: Parainfluenza virus 5.
HeLa cells: An immortal cervical cell line, isolated from Henrietta Lacks in 1952.
MRC-5 cells: Isolated from the normal lung tissue of a male embryo. MRC-5 has applications in viral vaccine development and efficacy testing.
We observed that the PIV-5 V and Middle East respiratory syndrome coronavirus (MERS-CoV) ORF4a proteins enhance protein expression 100- to 500-fold in vitro in IFN-competent HeLa and MRC5 cells.
We found that the MERS-CoV ORF4a protein partially abates dose nonlinearity in vivo, and that ruxolitinib, a potent Janus kinase (JAK)/signal transducer and activator of transcription (STAT) inhibitor, but not the IIPs, enhances protein expression of saRNA in vivo.
Both the PIV-5 V and MERS-CoV ORF4a proteins were found to enhance the percentage of resident cells in human skin explants expressing saRNA and completely rescued dose nonlinearity of saRNA.
Finally, we observed that the MERS-CoV ORF4a increased the rabies virus (RABV)-specific immunoglobulin G (IgG) titer and neutralization half-maximal inhibitory concentration (IC50) by ∼10-fold in rabbits, but not in mice or rats.
Express-as-you-go from the same saRNA template:
These experiments provide a proof of concept that IIPs can be directly encoded into saRNA vectors and effectively abate the nonlinear dose dependency and enhance immunogenicity.
They acknowledge that activation of interferon pathways (which you normally need to prime the immune system and destroy cancer cells) also inhibits expression of their desired protein:
Self-amplifying RNA is a cutting-edge vaccine platform but is known to activate interferons; this activation can augment immunogenicity but also inhibit translation of the antigen. Blakney et al. found that a cis-encoded innate inhibiting protein, MERS-CoV ORF4a, enhances both protein expression and immunogenicity of an saRNA rabies vaccine.
They weren’t considering SARS-CoV-2 when this paper was written in 2020, but the findings are directly applicable:
Self-amplifying RNA (saRNA) is a highly advantageous platform for both nucleic acid vaccines and therapeutics. Derived from an alphavirus genome,1 saRNA encodes the alphaviral replicase and a gene of interest (GOI), which replaces the structural proteins of the virus.
A variety of GOIs have been incorporated and shown to be highly immunogenic with the saRNA platform, including vaccine antigens for influenza,2 HIV-1,3,4 respiratory syncytial virus (RSV),3,5 and Ebola.6 Furthermore, saRNA vaccines are rapidly scalable, as they require a minimal dose compared to messenger RNA (mRNA),7 which is highly useful in the context of both normal vaccine production but also global pandemics.
saRNA is self-adjuvanting,8 as it activates a type I interferon (IFN) through endosomal sensing via Toll-like receptor (TLR)3, TLR7, and TLR8 as well as cytosolic sensing via melanoma differentiation-associated protein 5 (MDA5), retinoic acid-inducible gene I (RIG-I), protein kinase R (PKR), 2′-5′-oligoadenylate synthetase (OAS), as well as other possibly unknown pathways.9
As with mRNA tech, this is a problem, only more so:
While self-adjuvantation is advantageous and enhances the immunogenicity of RNA vaccines, this phenomenon is a double-edged sword, as innate recognition of mRNA upregulates the expression and activation of PKR and OAS, which leads to the inhibition of translation10 and degradation of cellular mRNA.11
Increasing doses of RNA are correlated with an increase of injection site reactions as well as systemic adverse events in human patients,12 which are likely due to the innate immune response to RNA.
They may work in the lab, but not in humans:
Furthermore, there is a disparity between the immunogenicity of RNA vaccines in preclinical animal models and human clinical trials, wherein RNA formulations are highly potent in lower order animal species such as mice, ferrets, and even nonhuman primates, but exhibit orders of magnitude lower potency in humans.12
This is likely due to inherent differences in innate immunity between different species, such as transcriptionally diverging genes that encode cytokines and chemokines.13
Double-stranded RNA (dsRNA) form the genetic material of some viruses, is highly immunogenic and play a role in the activation of the innate immune system. They can also enhance the effects of tumour immunotherapy and inhibit metastasis5.
Unfortunately, as a by-product, it is also associated with saRNA vaccine failure and side effects:
Innate immune sensing is especially critical for saRNA, as the self-replication leads to an exponential increase in copies of RNA in the cytoplasm as well as double-stranded RNA (dsRNA) intermediates.14,15
One potential strategy to reduce detrimental effects of type I IFN activation on RNA vaccines is to encode innate inhibiting proteins (IIPs) directly in the RNA, similar to the mechanism by which RNA viruses evade innate immune recognition and dampen the IFN response.16
What could possibly go wrong?
The previous study from 2017 also focused on these proteins:
Liu et al.17 observed that co-transfection of the E3, K3, and B18R (EKB) protein of vaccinia virus and non-structural protein 1 (NS1) of influenza A virus enhanced translation of mRNA.
Beissert et al.18 similarly observed enhanced protein expression in vitro and in vivo with the EKB proteins encoded by mRNA. However, this approach requires administration of two different types of RNA and does not guarantee co-localization of the IIP in the same cell as the GOI mRNA, which is imperative for overcoming innate sensing.
They screened a rogue’s gallery of viruses to find the best proteins to suppress our innate immune responses after vaccination:
In this study, we screened a library of saRNA constructs with cis-encoded IIPs and determined the effect on protein expression and immunogenicity. We chose a range of IIPs based on their varied targets in the type I IFN pathway (Figure 1; Table 1), including the herpes simplex virus 2 (HSV-2) US1,19 herpes simplex virus 1 (HSV-1) US1 and US11,20,21 Orf virus OV20.0L,22,23 bovine viral diarrhea virus (BVDV) Npro,24,25 parainfluenza virus 5 (PIV-5) V,26,27 Middle East respiratory syndrome coronavirus (MERS CoV) M,28, 29, 30 and ORF4a,31,32 Langat virus NS5,33,34 and influenza virus NS1 proteins.35
They cross-check against the human cell lines discussed:
We characterized the library of saRNA IIP constructs in vitro in mouse, rabbit, nonhuman primate, and human cells with varying IFN competencies, as well as in vivo for both intracellular and secreted protein expression.
Ruxolitinib is a targeted cancer growth blocker that is used to treat myeloproliferative neoplasms, such as myelofibrosis and polycythaemia vera. It is also used to treat patients with IgG4-RD.
Furthermore, we characterized how co-formulation with ruxolitinib, a potent US Food and Drug Administration (FDA)-approved Janus kinase (JAK)/signal transducer and activator of transcription (STAT) inhibitor,36 affects protein expression in vivo in mice and ex vivo in human skin explants.
We characterized the immunogenicity of IIP saRNA encoding the rabies virus (RABV) glycoprotein in mice, rats, and rabbits, including antibody titers and viral neutralization.
Finally, we characterized activation of IFN regulatory factor 3 (IRF3) and nuclear factor κB (NF-κB) transcription factors and cytokine responses in response to wild-type (WT) and IIP saRNA.
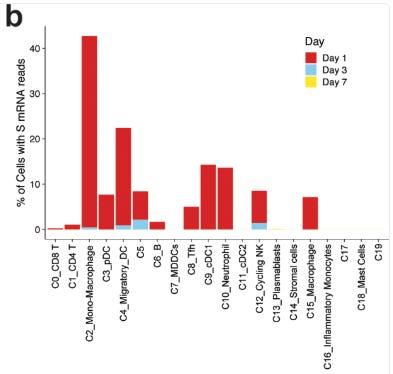
One of the rebuttals I had regarding our m1u paper was that it wasn’t a problem as it only suppressed immunity in the transfected cells. But it is something of a moot point if 50-60% of your migratory macrophages and dendritic cells are affected, as this study from 2020 demonstrated:
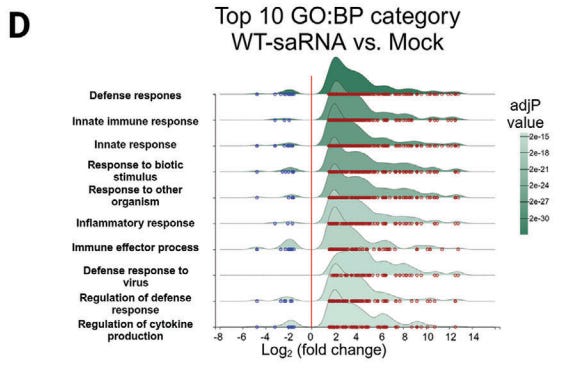
Quantification of cells with at least one read per cell demonstrated that 40% of monocyte/macrophages and 20% of migratory DCs contained spike mRNA on day 1 (Fig. 6b). The frequency of these cells increased after vaccination (Fig. 6c).
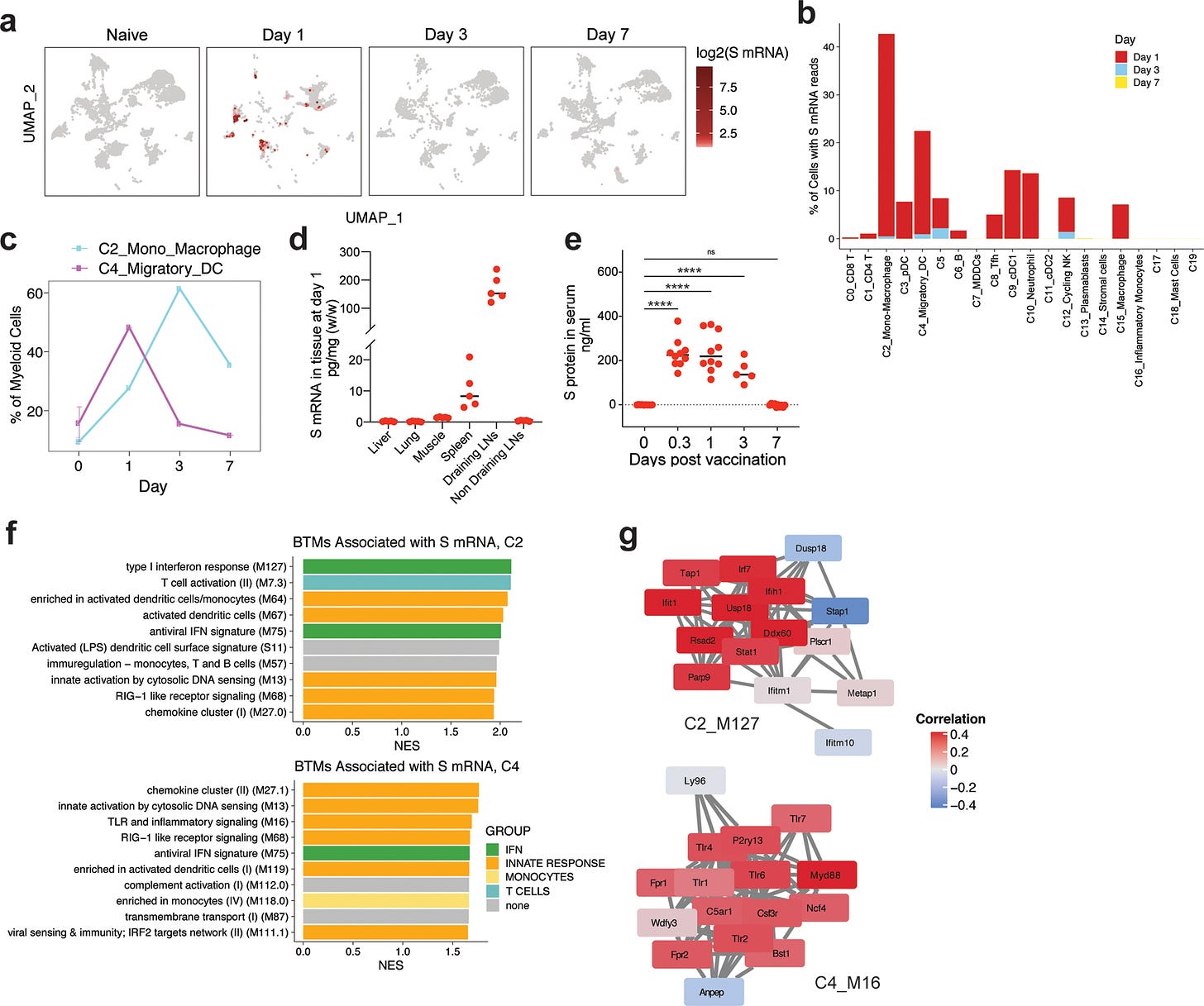
From: “Mechanisms of innate and adaptive immunity to the Pfizer-BioNTech BNT162b2 vaccine” (2020)
As we observed that the IIPs exhibit differences in protein expression depending on the species of the cell type in vitro, we sought to test the saRNA IIP constructs in a more clinically relevant human skin explant model.
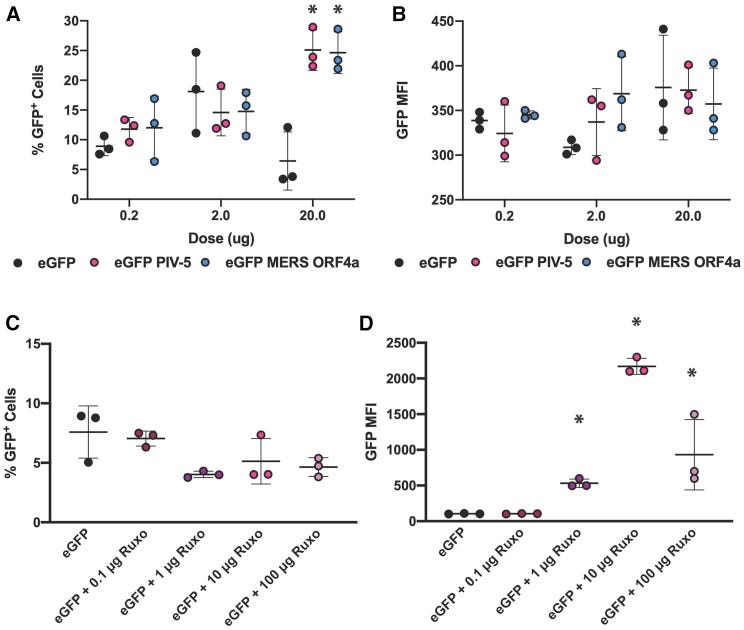
We characterized both the quantity (% of enhanced green fluorescent protein [EGFP]+ cells) and the quality of protein expression (median EGFP fluorescence intensity per cell) in resident human skin cells with incorporations of the PIV-5 V and MERS-CoV ORF4a proteins, as well as co-formulation with ruxolitinib (Figure 4).
We tested doses of 0.2, 2, and 20 μg of the EGFP saRNA with the PIV-5 V and MERS-CoV ORF4a proteins (Figures 4A and 4B) and observed that increasing the dose of the WT construct from 0.2 to 2 μg resulted in an increase of the percentage of EGFP+ cells from 10% to 18%, but when the dose was increased to 20 μg the percentage of EGFP+ cells plummeted to ∼5%.
However, for the PIV-5 and MERS-CoV constructs, there was a linear dose increase with increasing dose of saRNA.
This figure shows how expression of EGFP in human skin explants was significantly higher than the control when the viral inhibitory proteins were expressed.
Note the percentage of cells transfected by the saRNA construct at different doses in A. Even a dose as low as 0.2μg led to 7-17% of cells to express EGFP protein:
In some cell lines, this anti-cancer drug enhanced saRNA protein expression:
Next, we tested how incorporating doses of ruxolitinib, ranging from 0 to 100 μg, affected saRNA expression in human skin-resident cells. We observed that co-formulation of ruxolitinib with saRNA did not have any effect on the percentage of EGFP+ cells (Figure 4C), although there was a slight trend that increasing the dose of ruxolitinib actually decreased the percentage of EGFP+ cells from ∼8% to ∼5%.
EGFP: Enhanced Green Fluorescent Protein, which fluoresces strongly under blue or UV light.
However, we did observe a profound effect on the per-cell quality of EGFP expression (Figure 4D); increasing the dose of ruxolitinib increased the EGFP MFI from ∼100 to ∼2,000 at a 10-μg dose of ruxolitinib, although the MFI decreased to ∼1,000 with a 100-μg dose of ruxolitinib.
Similarly, to the cells expressing the saRNA PIV-5 and MERS-CoV ORF4a proteins, we found that ruxolitinib enhanced protein expression in the immune cells, as opposed to epithelial cells and fibroblasts, and specifically increased uptake in T cells, Langerhans cells, leukocytes, and NK cells (Figures S13A and S13B).
Taken together, these data show that the IIP replicons enhance expression in immune cells by increasing the percentage of cells expressing saRNA, while ruxolitinib enhances protein expression on a per cell basis.
They then demonstrated that MERS-CoV ORF4a and PIV-5 V proteins downregulate IFN signalling pathways, including NF-κB and IRF3:
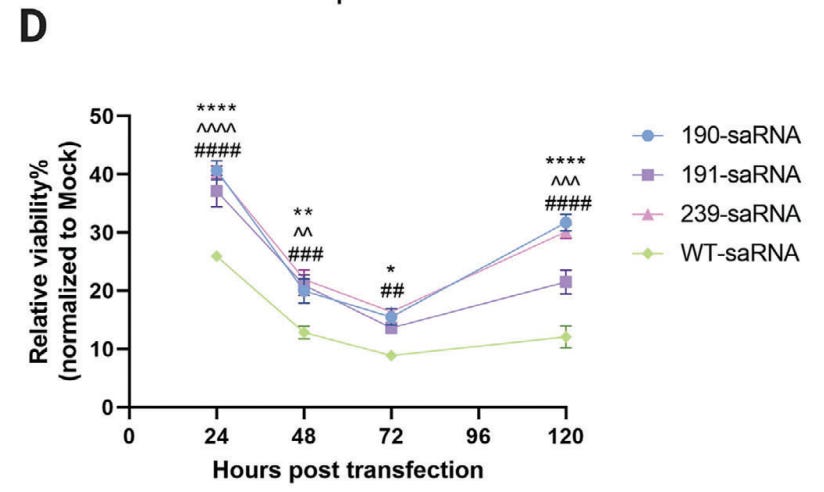
We observed that active NF-κB was upregulated after 4 h in samples that were treated with WT fLuc saRNA or tumor necrosis factor (TNF)-α (positive control), which both had an optical density (OD)450 of ∼2.2, whereas the PIV-5 and ORF4a constructs significantly downregulated the quantity of active NF-κB to OD450 of 0.42 and 0.52 (p = 0.0049 and 0.0031), respectively.
These data indicate that the PIV-5 V and MERS-CoV ORF4a proteins inhibit IFN activation in response to saRNA by downregulating NF-κB and IRF3 activity.
The “T” at the end of the dashed lines denotes blocking of that part of the signalling pathway. In this case both PIV-5V and ORF4a work together to turn off the cells innate immunity, so as to promote the preferred immune pathway via expression of the target protein.
I.e, as per the subtitle: “in order to build your immunity, we need to destroy it”:
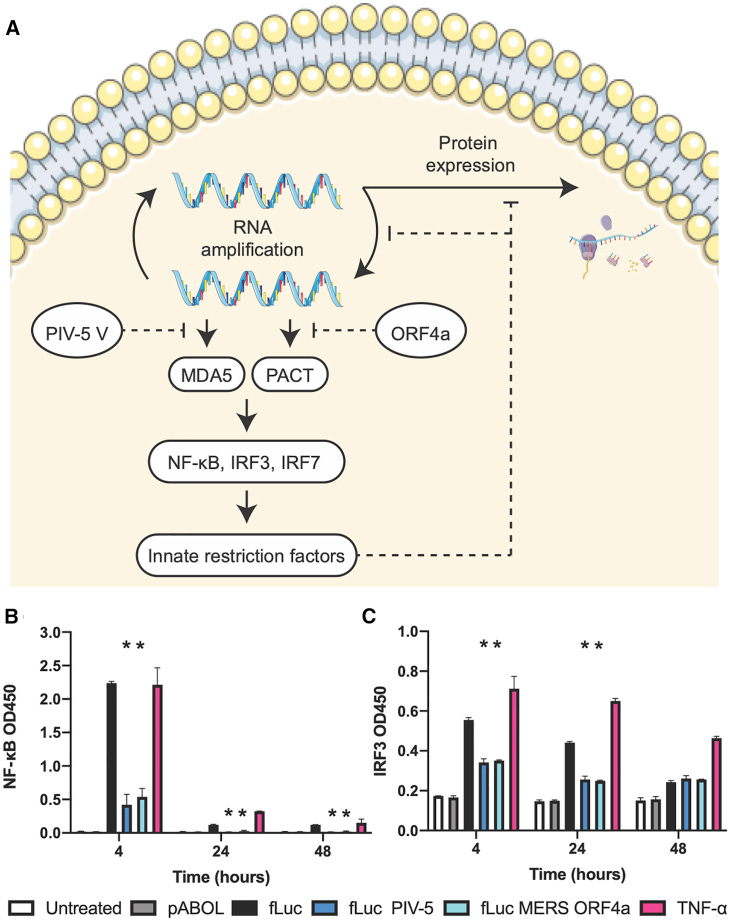
The protein designs, cells, and mutations characterized in these experiments offer insights into the mechanism by which the PIV-5 V and MERS-CoV ORF4a proteins increase protein expression (Figure 6).
MDA5: Melanoma differentiation-associated protein 5, a cytosolic sensor that recognizes viral double-strand RNA and then triggers the transcription of genes encoding type I IFN.
The PIV-5 V protein blocks MDA5 and IRF3 by binding to MDA5,26,27 whereas the MERS-CoV ORF4a protein binds to dsRNA and suppresses protein activator of the IFN-induced protein kinase (PACT) triggering of MDA5 and RIG-I.29, 30, 31
We observed that the PIV-5 V and ORF4a proteins downregulate IRF3 and NF-κB activation as compared to WT saRNA (Figures 6B and 6C).
This is similar to previous studies, which show that MERS-CoV accessory proteins interfere with innate antiviral signaling pathways, including the NF-κB-mediated response.47
A high degree of immune suppression occurs in humans, even at low levels of translation. Its also worth noting that in Part 1 Kostaive took 31 days to become undetectable in plasma, but it was still active in muscle and lymph nodes.
It should, of course, be contraindicated for cancer patients and the immune suppressed, i.e. the populations who are considered high priority for vaccination.
This is absurd, but nothing new:
While we observed minor differences in the mouse and human PBMC response to transfection with WT, PIV-5 V, and MERs-CoV ORF4a saRNA, this is likely due to low levels of transfection using pABOL (Figures S15 and S16). However, these results indicate that despite low levels of translation, the IIPs impact the cytokine profile of PBMCs and affect different cytokines depending on the species.
In conclusion:
To our knowledge, we are the first to observe that the IIPs enhance the percentage of cells expressing saRNA, whereas ruxolitinib enhanced the expression per cell.
A bit like being “a little bit pregnant”, there appears to be no such thing as a partial, or selective immune suppression:
We postulate that saRNA induces a binary state in a cell, wherein the replicase machinery enables maximal protein expression or the protein translation is massively inhibited by the IFN response.
A recurrent theme from the registration documents and papers like this is mechanisms remain incompletely understood, especially in humans, yet they are happy to “shoot from the hip” and go straight to commercial production and rollout:
We have previously observed that the delivery system can be tailored to increase the percentage of cells expressing a reporter protein in a similar manner to the IIPs,2,51, 52, 53, 54 but the mechanism behind this is not well understood and warrants further studies.
Given these promising results, we postulate that the MERS-CoV ORF4a protein may enhance immunogenicity of saRNA vaccines in humans, and it may also be useful for saRNA application to protein replacement therapies,55,56 although clinical utility will require evaluation of any anti-vector immune response to the replicase or IIPs.
These experiments provide a proof of concept that IIPs can be directly encoded into saRNA vectors and effectively abate the nonlinear dose dependency and enhance immunogenicity.
One limitation of this current study is that neither preclinical animal models nor human skin explants completely recapitulate the entire human immune system, which is the ultimate target of this work.
As indicated by the mechanistic studies, different aspects of the IFN pathway can be targeted and increase saRNA expression, thus motivating probing of combinations of IIPs and other IFN inhibitions strategies, such as ruxolitinib.
While these results are currently limited to preclinical animal models and human skin explants, future studies are warranted to study how these vectors enhance effectiveness of saRNA in human clinical trials.
In vitro adaptive passaging
Our final paper is from 2024 and discusses how serial passage of saRNA may be used to select alphavirus mutations that are favourable for reducing immunogenicity still further and for enhancing protein expression as a consequence.
Key takes from “A Novel Self‐Amplifying mRNA with Decreased Cytotoxicity and Enhanced Protein Expression by Macrodomain Mutations” by Gong et al.6:
The efficacy and safety of self‐amplifying mRNA (saRNA) have been demonstrated in COVID‐19 vaccine applications. Unlike conventional non‐replicating mRNA (nrmRNA), saRNA offers a key advantage: its self‐replication mechanism fosters efficient expression of the encoded protein, leading to substantial dose savings during administration.
Consequently, there is a growing interest in further optimizing the expression efficiency of saRNA. In this study, in vitro adaptive passaging of saRNA is conducted under exogenous interferon pressure, which revealed several mutations in the nonstructural protein (NSP).
Notably, two stable mutations, Q48P and I113F, situated in the NSP3 macrodomain (MD), attenuated its mono adenosine diphosphate ribose (MAR) hydrolysis activity and exhibited decreased replication but increased payload expression compared to wild‐type saRNA (wt saRNA).
Transcriptome sequencing analysis unveils diminished activation of the double‐stranded RNA (dsRNA) sensor and, consequently, a significantly reduced innate immune response compared to wt saRNA.
Furthermore, the mutant saRNA demonstrated less translation inhibition and cell apoptosis than wt saRNA, culminating in higher protein expression both in vitro and in vivo.
These findings underscore the potential of reducing saRNA replication‐dependent dsRNA‐induced innate immune responses through genetic modification as a valuable strategy for optimizing saRNA, enhancing payload translation efficiency, and mitigating saRNA cytotoxicity.
This research shows an example that reducing innate immunogenicity to increase payload protein expression of self‐amplifying RNA via a single mutation in the macrodomain. The mutant saRNA attenuates the activation of the double‐stranded RNA sensor as well as the downstream interferon response.
It persists in your tissues for longer, and “reduced cytotoxicity” is meant to mean less innate immunity associated adverse events. However, this doesn’t do anything to reduce the cytotoxicity of the LNP carrier platform or of the target protein, such as Spike:
It has improved ribosomal RNA stability and reduced cytotoxicity, leading to enhanced payload protein expression.
The “turtles all the way down” circular argument:
Overall, the structure of saRNA is designed to enable efficient replication and expression of the target proteins, making it a promising platform for vaccine development and other therapeutic applications.[ 9 , 10 ] The approval of a COVID‐19 saRNA vaccine in Japan in 2023 further validates its safety profile.[ 11 ]
In comparison to non‐replicating mRNA (nrmRNA), a significant advantage of saRNA lies in its higher and more sustained expression of exogenous genes, leading to a reduction in administered doses.
The registration documents showed that, in spite of this, they still want you to have as many boosters of Kostaive as possible.
Why?
Willie Sutton’s Law: “Its where the money is”.
As for safety, when this paper was published in 2024, Kostaive had been rolled out commercially and yet these points weren’t ever addressed satisfactorily in the submission documents:
Meanwhile, the replicative nature of saRNA also raises potential safety concerns, as uncontrolled replication could lead to cellular toxicity or immune‐mediated adverse reactions.[ 12 , 13 ] Therefore, optimizing saRNA sequences to further enhance expression efficiency and safety is still a focal point.
The challenge, solved by using N1-methyl-pseudouridine (m1Ψ) in mRNA COVID vaccines :
The utilization of nucleotide modifications to evade recognition by pattern recognition receptors (PRRs) has been highly effective in reducing the innate immune response and improving gene expression efficiency in nrmRNA.[ 14 , 15 ]
Not so fast. m1u doesn’t work effectively in saRNAs, as it doesn’t amplify:
However, incorporating such modifications into saRNAs presents unique challenges due to the alphavirus‐derived sequences they contain. Nucleotide modification in saRNAs is often hindered by the risk of compromising the functionality of the replication machinery, thus posing a significant challenge.
Furthermore, even a nucleotide modified saRNA can be viable, many copies of progeny RNA of amplification product are unmodified.[ 16 , 17 ]
Contaminants are intrinsic to these products, and its a huge challenge. I discuss this further in part 3:
In addition to unmodified nucleotides, replication byproducts including double‐stranded RNA (dsRNA) intermediates, cap0‐structured progeny RNA, and capless 5′ triphosphate RNA are all recognized as pathogen‐associated molecular patterns (PAMPs) by PRRs.
Upon recognition, these patterns trigger antiviral immune responses, including but not limited to the interferon (IFN) or nuclear factor kappa‐light‐chain‐enhancer of activated B cells (NF‐κB) signaling pathways, inducing abundant interferon to stimulate genes such as protein kinase R (PKR), 2′‐5′‐oligoadenylate synthetase (OAS) and interferon induced proteins with tetratricopeptide repeats (IFIT) to directly restrict saRNA replication and expression.
Pepini et al. observed a significant increase in the expression of saRNA in mice lacking type I interferon receptors or in naïve mice whose type I interferon receptors were blocked by antibodies.[ 18 ]
Furthermore, Zhong et al. demonstrated the efficient suppression of the type I IFN response and enhanced translation of saRNA through the topical application of innate immune inhibitors.[ 19 ]
Hold it there: Topical application of innate immune inhibitors could suppress your type I IFN innate immunity signalling?
Let’s check out ref. [19]:
Clobetasol: A corticosteroid applied to the skin as a cream, foam, gel, liquid, solution, ointment, or shampoo.
I discussed the many risks to taking steroids at high doses or for long durations here, and clobetasol also stopped the target antibodies being formed.
They also used cellulose to remove as much dsRNA as possible. However, this technique isn’t currently commercially viable:
“Current methods for dsRNA elimination use either high-performance liquid chromatography or microcrystalline cellulose, rendering the process complex, expensive, toxic, and/or time-consuming.7”
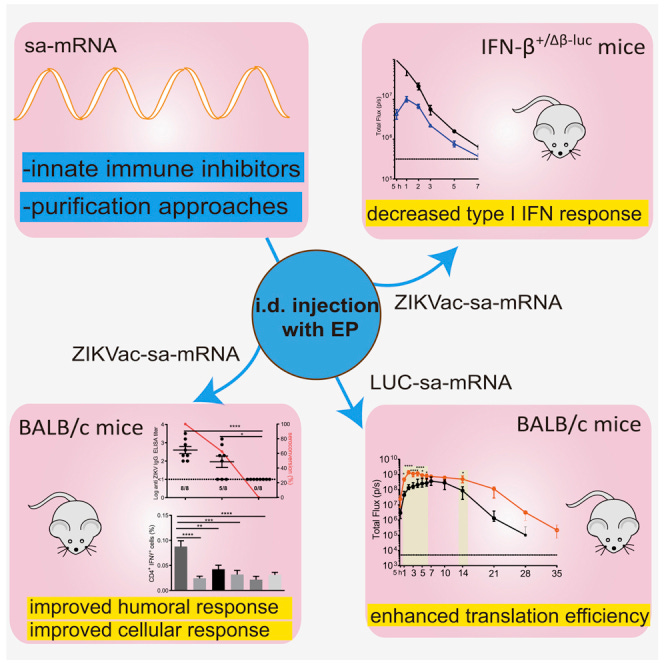
Among the investigated inhibitors, we found that corticosteroids and especially topical application of clobetasol at the sa-mRNA injection site was the most efficient in suppressing the type I IFN response and increasing the translation of sa-mRNA.
However, clobetasol prevented formation of antibodies against sa-mRNA-encoded antigens and should therefore be avoided in a vaccination context.
Residual dsRNA by-products of the in vitro transcription reaction are known inducers of immediate type I IFN responses.
We additionally demonstrate a drastic reduction of these dsRNA by-products upon cellulose-based purification, reducing the innate immune response and improving sa-mRNA vaccination efficacy.
From: “Corticosteroids and cellulose purification improve, respectively, the in vivo translation and vaccination efficacy of sa-mRNAs“ (2021)
These findings indicate that an exaggerated innate immune response significantly inhibits the expression levels and sustained expression of payload. Hence, improved saRNA expression efficiency might be accomplished by suppressing the IFN response.
The original sin of engineered RNA products - there can be no safe, effective solution:
Several strategies have been reported to improve saRNA expression. One strategy introduces viral innate inhibiting proteins, either by encoding simultaneously with payload or administered concomitantly with saRNA to evade the immune response.[ 20 , 21 ]
Nevertheless, the potential safety issues associated with both the virus protein itself and the robust suppression of the natural immune response require careful and thorough evaluation.
Another strategy entails the introduction of alphavirus translation enhancers to boost subgenomic RNA replication or evade host translation inhibition.[ 22 , 23 ]
In other words, we don’t know if this would work in humans:
However, potential affection for protein functionality resulting from the fusion of enhancing sequences with payload requires further confirmation.
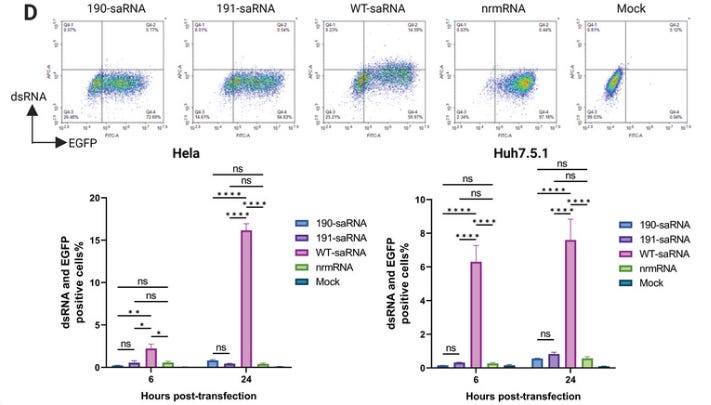
On cytotoxicity. Wild type saRNA left only 10% of transfected Huh7.5.1 cells viable (i.e. alive and functional) by 72 hours. This is potentially happening to any transfected cells, including immune cells, ovaries, testes, heart, liver, kidneys, cardiac, pancreatic, immune cells etc.
Remember, the process is mimicking the cytotoxicity of an alphavirus to some degree. Its no surprise that lethargy is one of the most reported AEs, along with lymphopenia:
Lastly, we assessed the viability of cells transfected with mutant saRNAs. Despite a notable reduction in cell viability observed in all saRNA‐transfected cells, the three mutants exhibited significantly lower cytotoxicity compared to the wt saRNA across all tested time points (Figure 2D). The cell viability of mutant 191 slightly decreased compared to the other two mutant saRNAs after 5 days of transfection.
They measured dsRNA contamination levels. In Hela cells over 16% of cells transfected with wt saRNA tested positive after 24 hours, and ~8% of Huh7.5.1 cells (Source tissue: Hepatocellular carcinoma).
nrmRNA: Conventional non‐replicating m1u-modified mRNA.
2.3.3. Reduced dsRNA Level
Since replication of saRNA produces high amounts of dsRNA, which triggers strong innate immune responses in the transfected cells, we measured dsRNA levels in the transfected cells. Consistent with reduced replication, mutants 190 and 191 exhibited much lower dsRNA levels in both HeLa and Huh7.5.1 cells, showing levels similar to the nrmRNA group (Figure 3D; Figure S3, Supporting Information).
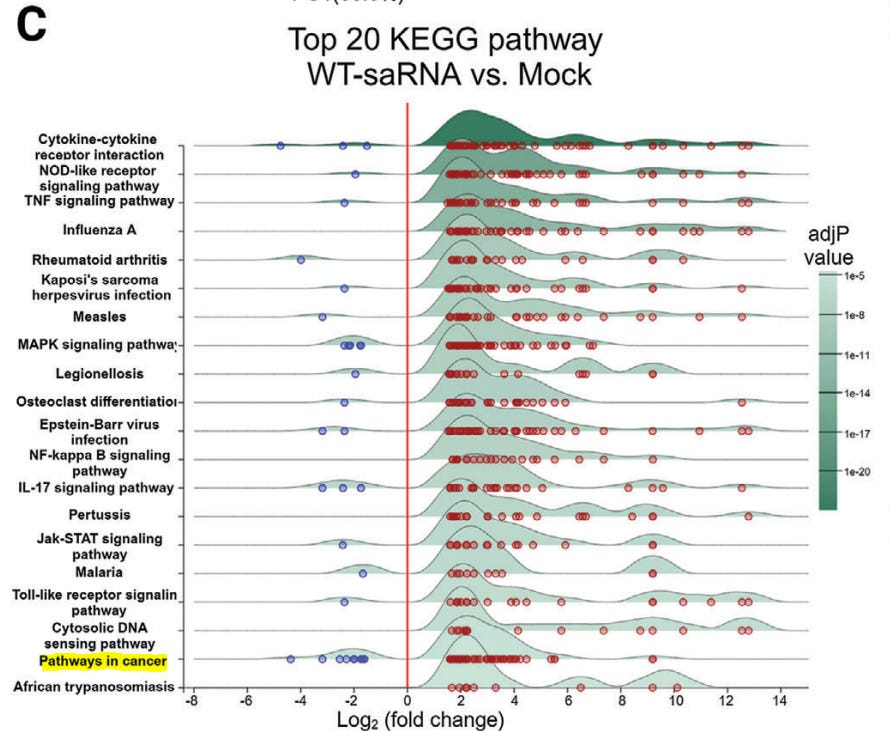
Of interest, they performed a Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of differentially expressed genes. These are the genes they need to suppress in order to increase target protein expression and duration. Note that in doing this your innate immunity non-target infections is significantly diminished, along with “Pathways in cancer” - the focus of this series.
2.4 The Mutant saRNAs Induce a Less Intrinsic Innate Immune Response
To gain insight into the changes in cellular activities following transfection with saRNA, we extracted total cellular RNA from HeLa cells transfected with mutant 190, 191, wt saRNAs, and also nrmRNA, respectively, for transcriptome analysis.
Principal component analysis (PCA) revealed that compared to mock-transfected cells, the nrmRNA induced minimal transcriptomic changes, while wt saRNA induced significant transcriptome disparity, far exceeding the changes observed in cells transfected with mutants 190 and 191 (Figure 4A).
Extracts from Figure 4:

Note the highly successful inhibition of immune signalling pathways of conventional m1u-modified nrmRNA.
From our review paper, this shows how m1u fails to suppress metastasis of melanoma cancer cells to the lungs of mice.
PR8HA-LNP is an unrelated influenza H1N1-derived antigen, also called A/PoRico/8/1934.
In contrast, egg-derived ovalbumin (OVA-LNP) was far more immunogenic when its signalling was not suppressed:
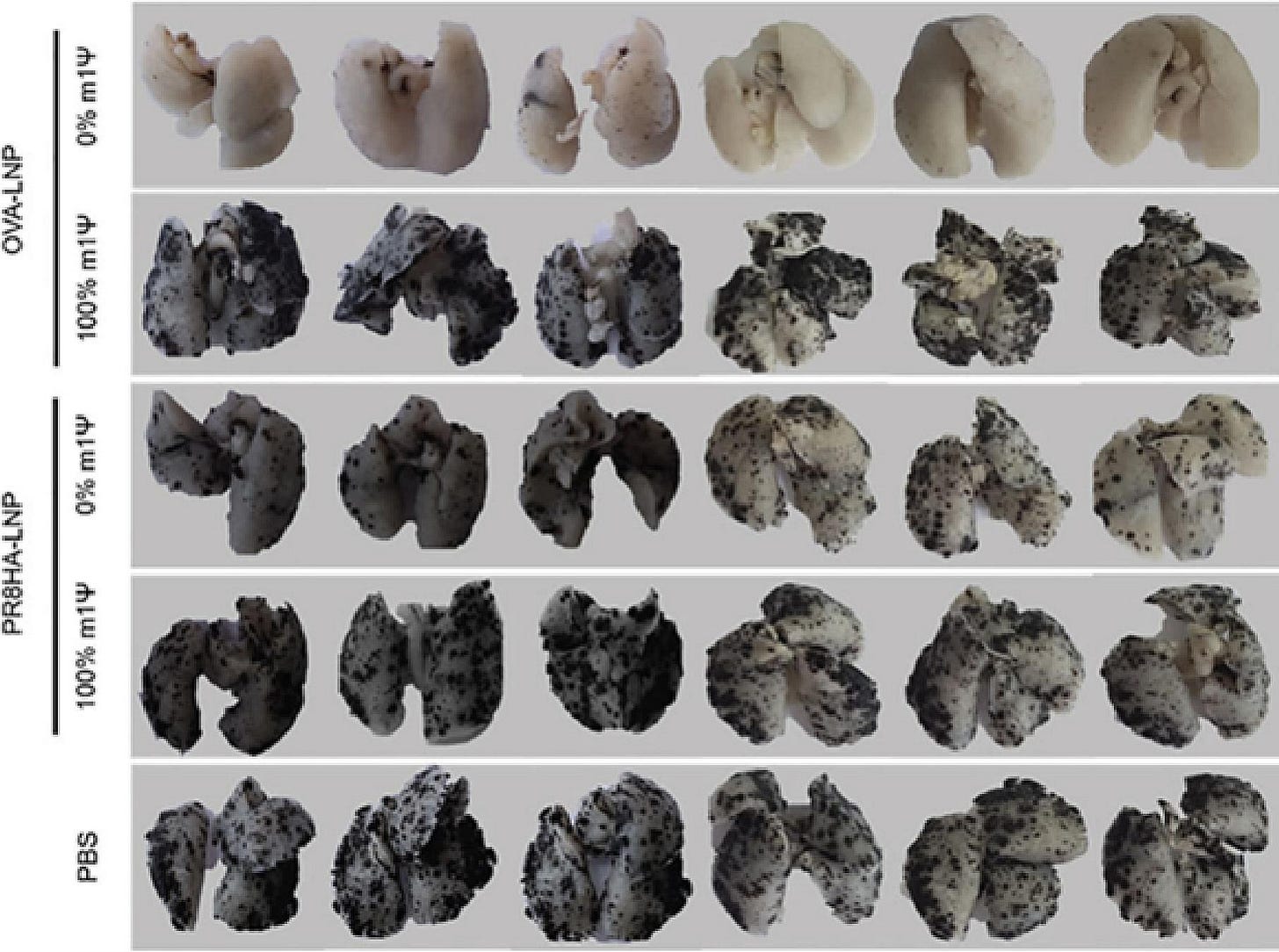
From: “Review: N1-methyl-pseudouridine (m1Ψ): Friend or foe of cancer?” (2024)
Concluding remarks
The final part of the trilogy will discuss the involvement of some of the above IIPs in cancer pathways, a potential therapeutic to mitigate some of the effects, the importance of innate immune responses to cancer cells, more on the consequences of the commercially unsolvable dsRNA contamination nightmare, and the saRNA patent landscape.

From our review paper’s discussion, a cautionary warning.
Since writing this, many medical centres and oncologists are reported to be extremely busy, with long waiting lists to see a consultant:
… The main limitation of this review is that, to date, there are few reports associating the use of m1Ψ in mRNA vaccines with cancer development, making the relationship between cause and effect difficult to determine. Precisely for this reason, it is urgent to carry out long-term studies in the animal model to verify or refute such a possibility, especially given the long asymptomatic latency periods typical of many cancers. In the analysis by Nadler & Zurbenko [146], 35 of 44 patients, representing 89 % of cases, had cancers that progressed without detection for 10 years or longer.
From: “Review: N1-methyl-pseudouridine (m1Ψ): Friend or foe of cancer?” (2024)
https://pubmed.ncbi.nlm.nih.gov/38583833/
Full paper:
Part 3:
“Anything but the Thing…”
And finally…

References
‘Glossary: Standard IARC Classification’. Accessed 13 December 2024. https://ec.europa.eu/health/scientific_committees/opinions_layman/en/electromagnetic-fields/glossary/ghi/iarc-classification.htm.
Rubio-Casillas, Alberto, David Cowley, Mikolaj Raszek, Vladimir N. Uversky, and Elrashdy M. Redwan. ‘Review: N1-Methyl-Pseudouridine (m1Ψ): Friend or Foe of Cancer?’ International Journal of Biological Macromolecules 267, no. Pt 1 (5 April 2024): 131427. https://doi.org/10.1016/j.ijbiomac.2024.131427.
Beissert, Tim, Lars Koste, Mario Perkovic, Kerstin C. Walzer, Stephanie Erbar, Abderraouf Selmi, Mustafa Diken, Sebastian Kreiter, Özlem Türeci, and Ugur Sahin. ‘Improvement of In Vivo Expression of Genes Delivered by Self-Amplifying RNA Using Vaccinia Virus Immune Evasion Proteins’. Human Gene Therapy 28, no. 12 (1 December 2017): 1138–46. https://doi.org/10.1089/hum.2017.121.
Blakney, Anna K., Paul F. McKay, Clément R. Bouton, Kai Hu, Karnyart Samnuan, and Robin J. Shattock. ‘Innate Inhibiting Proteins Enhance Expression and Immunogenicity of Self-Amplifying RNA’. Molecular Therapy 29, no. 3 (3 March 2021): 1174–85. https://doi.org/10.1016/j.ymthe.2020.11.011.
Yu, Yulin, Yinmei Tian, Yang Li, Xianya Qin, Xiaonan Li, Qian Hu, Chuansheng Fu, et al. ‘Self-Generated Double-Stranded RNA for Enhancing Tumor Immunotherapy and Metastasis Inhibition’. Nano Today 55 (1 April 2024): 102173. https://doi.org/10.1016/j.nantod.2024.102173.
Gong, Yue, Danni Yong, Gensheng Liu, Jiang Xu, Jun Ding, and William Jia. ‘A Novel Self-Amplifying mRNA with Decreased Cytotoxicity and Enhanced Protein Expression by Macrodomain Mutations’. Advanced Science 11, no. 43 (2024): 2402936. https://doi.org/10.1002/advs.202402936.
Natural Wood‐Derived Macroporous Cellulose for Highly Efficient and Ultrafast Elimination of Double‐Stranded RNA from In Vitro‐Transcribed mRNA - Yuan - 2024 - Advanced Materials - Wiley Online Library. Accessed January 10, 2025. https://onlinelibrary.wiley.com/doi/full/10.1002/adma.202303321?saml_referrer











Before I read your novella I have to say the introduction was brilliant, it was funny all the way through but sets the scene for our potential future. If they could they would jab every single human on the planet and yes even our pet gerbils are not safe.
"This may be referred to as the “original sin” of the mRNA platform: in order to express enough antigens for long enough to generate an immune response it must, by necessity, suppress innate immune responses to itself. "
The irony is it suppresses the immune response to the very protein it's supposed to induce protection against. It's so batshit it's almost impossible to conjure up an analogy fitting; sir, please remove the body armour so your body can experience the full effects of these gunshots, then you can put the body armour back on.